NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Institute of Medicine (US) Forum on Emerging Infections; Knobler SL, Lemon SM, Najafi M, et al., editors. The Resistance Phenomenon in Microbes and Infectious Disease Vectors: Implications for Human Health and Strategies for Containment: Workshop Summary. Washington (DC): National Academies Press (US); 2003.

The Resistance Phenomenon in Microbes and Infectious Disease Vectors: Implications for Human Health and Strategies for Containment: Workshop Summary.
Show detailsMICROBIAL RESISTANCE: ADOPTING AN EVOLUTIONARY APPROACH
Resistance of infectious microorganisms to therapeutics is a serious—and growing—health threat in the developed and developing world alike. Introductions of new classes of antimicrobial drugs have been followed, often quickly, by the emergence of resistant microorganisms. Drug-resistant pathogens are now commonplace in hospitals and other health care settings, and they are increasingly found in communities as well. Compounding the problem, drug discovery efforts are yielding fewer truly new leads toward novel classes of antimicrobial agents.
In addition to the direct effect of antimicrobial resistance on human health, there also is an economic cost. In one estimate, for example, the hundreds of thousands of penicillin-resistant and methicillin-resistant infections that occur in the United States each year add roughly $30 billion to the national health care bill. Higher costs that result from antimicrobial resistance are particularly burdensome for developing nations, and in some cases effective therapies are now priced out of practical reach.
One major factor contributing to the emergence of drug-resistant microbes is the increase in the sheer volume of antimicrobial agents, particularly antibiotics, being used today. By one analysis, between 35 million and 50 million pounds of antibiotics are produced annually in the United States alone, for medical use in humans as well as agricultural use in a variety of animals and plants. Recent years also have seen an explosion of household products that contain antibacterial agents. This “antibacterial craze” defies the critical message that washing with soap and water is sufficient to provide hygiene to healthy individuals. Moreover, some studies indicate that bacteria emerging with resistance to these chemicals show decreased susceptibility to a number of antibiotics.
The unrelenting spread of antimicrobial resistance has been recognized for some time, with a number of public and private organizations issuing reports calling for action by the health care community, governments, and the public. The World Health Organization (WHO), for example, has declared antimicrobial resistance to be one of the top three issues in global health. Yet, efforts to manage antimicrobial resistance have, in general, remained insufficient in the face of the magnitude of the problem.
Not only are more—and better coordinated—efforts required. There also is a need to view antimicrobial resistance in a fundamentally different way. Where we once concentrated primarily on developing chemicals to eradicate pathogens, we now need to pay closer attention to the ecology and evolution of infection. Developing a fuller understanding of how microbes evolve when faced with drugs that threaten their survival may lead to innovative ways to bring them under control.
Indeed, genetics and evolutionary science hold some important lessons. Over the past 10 years, for example, scientists have expanded their knowledge of the breadth and complexity of the genetics of antimicrobial resistance. Hundreds of genes have been identified that render a variety of microbes resistant to a variety of drugs. These findings are encouraging laboratories, both public and private, to step up efforts to define new resistance genes and investigate their biochemical activity.
Evolutionary science has grown in sophistication and predictive power, and a body of theory and practical observation now exists that suggests possible strategies for helping to solve some of the problems that arise from the continued “arms races” between humans and microbes. For example, a number of ways have been demonstrated to slow down the evolution of drug resistance. These approaches include, among others, drug overkill strategies that reduce a microbe's potential for genetic mutation, direct observation therapy to ensure that patients complete their full courses of treatment, and alternating use of various antibiotic drugs to lessen the genetic pressure that promotes resistance. Another promising avenue beginning to be explored involves developing drugs that “engineer” a microbe's genetic mutations in a way that imposes a fitness cost and renders the pathogen less able to survive or evolve in its human host.
Use of such tactics follows from the realization that fighting infectious disease is only part of the health care battle. Suppressing the emergence of these diseases also should be an integral part of the agenda to foster the long-term promotion of human health.
ARMS RACES AND ANTIMICROBIAL RESISTANCE: USING EVOLUTIONARY SCIENCE TO SLOW EVOLUTION
Authors
Stephen R. Palumbi, Ph.D.1.Affiliations
A Thomson's gazelle can run like lightning late for a train: a fact that meshes beautifully with the prime place that this fleet animal holds on the menu of the world's fastest cat—the cheetah. The predator-prey drama unfolds with dinner on one side and death on the other, and the evolutionary dynamics that drive this system are very clear. Both cheetah and gazelle are in an arms race: where evolutionary change of the predator drives either compensatory change in the prey—or extinction. Any increase in gazelle speed or maneuverability exerts evolutionary pressure on the cheetah to notch up its own speed. The result of this arms race is a steady escalation of running ability, until both players challenge the maximum performance their basic construction will allow. Escalation caused by predator-prey arms races is a normal expectation of evolutionary models. Similar dynamics play out in many aspects of human-disease interactions, and similar evolutionary predictions have been validated many times.
Antibiotics exert a powerful evolutionary force, driving infectious bacteria to evolve powerful defenses against all but the most recently invented drugs. For example, when penicillin first became available in the middle 1940s, most infections caused by Staphylococcus aureus could be easily cured by a dose of 10,000 units (Neu, 1994). But by the late 1940s resistant strains were reported, and were so common by the early 1960s that hospitals in North America switched treatment strategies to the more powerful, and more expensive drug, methicillin (see Table 1-1). Methicillin resistance, discovered for the first time in Cairo in 1961, has spread so that by 2000, it rendered about 50 percent of the hospital-acquired infections of S. aureus resistant to the drug. Vancomycin, called the drug of last resort, cured these more powerful infections, but cases of resistance are increasing. In 1999, the FDA approved linezolid to treat vancomycin-resistant infections. The first linezolid-resistant bacteria are currently making the rounds.
TABLE 1-1
The History of Our Ongoing Race with Staphylococcus aureus.
This classic arms race between the evolutionary potential of bacteria and the industrial power of the modern pharmaceutical industry results in two important features of modern health care. First, antibiotic treatments are not static—they change over time as new strains of bacteria evolve and spread. Second, the substitution of treatments generally results in steady escalation of treatment costs. Generic amoxicillin is a standard treatment for childhood infections, with a cost of $4–$24. But so many strains of bacteria have evolved beta-lactamase enzymes that destroy the antibiotic that new formulations also include a beta-lactamase inhibitor, clavulanic acid. The resulting mixture, co-amoxyclav, has an augmented price of $64–$172 according to the Consumers Union of the United States (2001).
Overall these increases can have a substantial impact on health care costs. For example, if a hospital patient becomes infected with S. aureus, and the strain is resistant to penicillin, then the average hospital bill increases by about $9,000 in the United States (Abramson and Sexton, 1999). Should the strain prove to be resistant to methicillin as well, then the cost increase triples to $27,000. These costs add up to substantially increased expenditures in local hospitals: in the New York city area, costs of treating resistant S. aureus exceeded $400 million in 1999 (Rubin et al., 1999). The hundreds of thousands of penicillin-resistant and methicillin-resistant infections in the United States add to the national health care bill by an estimated $30 billion (Palumbi, 2001b).
Rapid Escalation of HIV
Probably one of the world's fastest evolving human diseases is also one of the clearest examples of arms races in health care and the escalation that results. Human immunodeficiency virus (HIV-1) has one of the fastest mutation rates on record due to a high error rate in its main genome-copying enzyme reverse transcriptase. The practical result of this high rate of mutation is that every infection of a new individual results in a unique evolutionary trajectory as the virus mutates into many slightly different forms. HIV is not a single infection, but a welter of different infections termed a quasi-species (Crandall, 1999).
This variation fuels rapid HIV evolution in response to two different sources of selection. This first source is the immune system of the person harboring the disease, which responds to HIV infection the way it should: by mounting a vigorous defense. But even though the immune system targets and destroys the first infective virus, by the time this defense is activated, the virus has mutated into so many forms that some of them evade immune detection. Mere weeks after the start of the immune response, amino acid evolution in the coat proteins that cover the viral shell has begun (see Figure 1-1). The cycle of immune targeting and viral escape repeats many times, each cycle representing a turning of the arms race wheel. Because the cellular hosts of HIV are T cells, which play an important role in the immune system, each arms race cycle reduces the strength of the immune system, eventually resulting in an immune collapse, and the onset of acquired immune deficiency syndrome (AIDS).
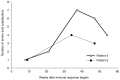
FIGURE 1-1
Rapid evolution of amino acid sequence of HIV coat proteins after the start of the immune response to infection. Within months, coat proteins evolve to evade immune detection. SOURCE: Wolinsky et al., 1996.
This is only the first of two major arms races with HIV. The second ignites when antiretroviral drugs are used to combat the virus. In every case, HIV evolves so quickly in response to this strong selection that antiretroviral drugs, used singly, have only a short window of effectiveness. For example, AZT was one of the first effective anti-HIV drugs. At first it worked well to reduce viral loads and to remove circulating viral RNAs from the bloodstream. This was because the reverse transcriptase enzyme used AZT instead of the normal DNA precursors when it copied viral RNA into DNA, and these altered DNA copies were non-functional (Palumbi, 2001a). But within 6–18 months, HIV levels rose in AZT-treated patients, and the virus no longer responded to the drug (see Figure 1-2). Studies conducted in vitro showed that HIV quickly evolved the ability to grow in the presence of AZT. Further studies showed that four amino acid substitutions were required to confer virtual immunity of HIV to AZT, and that these same four amino acid substitutions occurred independently in patient after patient (see Figure 1-3). Not only is this an excellent, experimental demonstration of convergent evolution, but it also shows the ability of selection and mutation to combine to produce rapid HIV evolution within each infected individual.

FIGURE 1-2
Evolution of AZT resistance in HIV occurs rapidly after the beginning of treatment. SOURCE: Larder and Kemp, 1989.
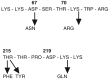
FIGURE 1-3
Four amino acid substitutions in the reverse transcriptase enzyme are sufficient to confer strong AZT resistance. These four substitutions occur in different order but accumulate in independent cases of the evolution of resistance. SOURCE: Larder and (more...)
HIV also evolves quickly in the presence of other drugs to become resistant. In fact there is no single drug to which HIV has not been able to evolve resistance. Ignoring this widespread and pervasive evolution would be a fatal mistake.
The global cost of this rapid evolution must also not be ignored, because HIV evolution demands huge payments in human lives and health care expenditures. AZT is relatively cheap to produce, and if not for evolution of resistance, we would have substantially overcome HIV years ago by deploying this and other anti-retrovirals. Instead, HIV evolution prevents us from using simple drug treatment strategies, and HIV sufferers in North America and Europe rely on triple-drug therapy to stall the progress of the disease. This treatment is expensive (about $15,000–$20,000 a year per patient in the United States in 2000 [Bozzette et al., 2001]), and contributes $6 billion–$8 billion to U.S. annual health care costs (Palumbi, 2001b). Such high costs are impossible to meet in many developing nations, especially those in Africa that are the epicenter of the global HIV epidemic. In this case, the escalation of costs during the arms race with HIV results in a disease which is only temporarily manageable medically and entirely unmanageable economically.
Escalation and Global Medical Care
Other human diseases are evolving so quickly that they too are more and more expensive to treat, causing grave concerns about the ability to treat them in poor populations. Tuberculosis (TB) is relatively easy to cure as long as a patient takes a long enough dose of the appropriate antibiotics. But because treatment demands a six-month drug course, it is common for people to cease taking the proper antibiotics before a cure is effected. Relapses after a partial drug course are common, and the strain of TB that re-emerges has a much higher chance of being resistant (Lewis, 1995). These resistant strains can take up to 200 days in a hospital to cure, at a cost that far exceeds the cost of curing the original, non-resistant strain. Likewise, malaria (Hastings and Mackinnon, 1998), leprosy (Honore and Cole, 1993), as well as a host of other bacteria like Enterococcus faecium (Schentag et al., 1998) have evolved strains that are expensive to treat. All these outcomes result from the same basic evolutionary mechanism of arms races and escalation.
Antibiotic resistance and other evolutionary responses to medical treatment parallel the evolution of resistance in insect pests, agricultural weeds, and fisheries populations under strong selective mortality (Palumbi, 2001b). Such evolutionary changes have long been heralded, but there has been little in the way of coordinated response to this threat. In some sense this lack of coordinated response stems from the failure of medical, agricultural, and public health industries to realize that insect and bacterial resistance are based on the same evolutionary principles, and therefore can be addressed in the same fundamental ways. Over the past few decades, evolutionary science has grown in sophistication and predictive power, and has produced a body of theory and practical observation that can be used to help solve some of the problems of arms race escalation outlined above. Certainly, this approach cannot invent new drugs or roll back resistance, but it can help in important ways.
Evolutionary Science and Arms Races
From the standpoint of evolutionary science, arms races can be understood as the outcome of antagonistic coevolution, where evolutionary change in one species induces change in a second that induces more change in the first. This positive feedback loop is common whenever one species causes mortality in a second, but it is not inevitable. Furthermore, the rate and the direction of coevolution can be changed. As a result, understanding the evolutionary machinery that drives arms races can provide clues about how to throw a wrench in the works. Over the last decade, a number of different ways to slow down, halt, or reverse arms races have been invented and tried. In some cases, these efforts have paid off well to reduce the problems of antibiotic resistance, and can provide some clues about efforts that may be valuable in the future.
Drug Overkill
If we are in an arms race with HIV and it is one of the most quickly evolving organisms known, why has there been recent victory over HIV? What stopped the HIV race? Current medical care in the United States emphasizes administration of triple drug therapy, the combined administration of reverse transcriptase inhibitors and protease inhibitors (Ho, 1995). This approach has reduced deaths due to HIV by several-fold since 1996, and it has greatly slowed the evolution of this disease within patients (Wong et al., 1997). But how?
Triple drug therapy works to slow evolution because it is a drug overkill strategy. The combination of drugs is so potent that even among the enormous numbers of viruses circulating in the bloodstream, none possesses the genetic mutations necessary to allow the virus to grow. Essentially, there is no variation among viruses for the ability to grow in the presence of triple drug therapy. Evolution is fueled by variation, and where there is none, evolution halts. As a result, in overkill strategies, not only are the viruses quelled by potent drugs, but their ability to evolve resistance defenses is seriously curtailed.
Unfortunately, this strategy for HIV has not effected any cures. It is sufficient to keep the virus in check and to prevent its rapid evolution, but it has not yet led to the cure of anyone with HIV. This means that our current drug strategy is best viewed as a temporary measure to slow down the HIV rampage while a more permanent solution is found.
Direct Observation Therapy
Hidden within the success of drug overkill strategies lies the reason for the failure of many less powerful antimicrobial approaches. If complete inhibition of viral growth explains slow HIV evolution during triple-drug treatment, then partial treatment can easily be seen as a major impetus to the evolution of resistance. This is one of the most important lessons that evolutionary biology can contribute to health management—that when it comes to the evolution of resistance diseases, half measures can kill.
To see why this is true, imagine that a population of bacteria in an infection varies in the susceptibility of individual cells to a drug, and that some cells die in the presence of a drug dose more quickly than others (see Figure 1-4). The median life span of infectious cells in the presence of the drug is about 6.5 days in this example. If treatment stops after this time, half the cells have died, but the other half survive, going on to reproduce. Assuming their offspring inherit their drug survival abilities, then the median life expectancy of the half that are left increases to about 9 days. A 10-day drug course eliminates 80 percent of the cells, but the survivors have a median drug resistance time of about 12 days.

FIGURE 1-4
Hypothetical relationship between survival of a population of infectious cells and the length of a drug treatment regimen. The graph and subsequent discussion assume wide, genetically based variation in susceptibility in the original population, and no (more...)
The increase in survival time among infectious cells after a drug treatment is called the “response to selection,” and it increases with the severity of the selection and the heritability of the trait in question. Only when the complete drug course is taken, and all the infectious cells in the body have been eliminated does the response to selection drop. At this point, selection is 100 percent, no cells are left to start a new generation (unless there is reinfection from outside), and evolution cannot proceed. Although this is a hypothetical example, and the response to a drug course may not be as smooth as depicted in Figure 1-4, the overall point is that the result of selection is always a more robust population, unless mortality is complete, or heritability is low.
These considerations show why there is an increasing call for rigorous adherence to antibiotic treatment protocols, and why partial doses of antibiotics are contributing so heavily to the evolution of drug resistance. Prolonged treatment with antimicrobials is also important because of their impact on commensal flora. Even judicious use of antimicrobials will not completely halt evolution of resistance because such evolution could also occur in normal bacterial flora that is exposed to antimicrobials during treatment of a pathogenic disease, or in pathogenic organisms inhabiting the outside environment. High titres of antimicrobials in soils or water, for example, may select for resistance (Ash et al., 2002). In management of tuberculosis, strict treatment protocols include six or more months of antibiotic use, largely because the slow-growing TB bacteria have a long drug survival time. To reduce the evolution of resistance caused by failure to complete such long courses, Direct Observation Therapy uses a protocol in which TB drugs are delivered daily to outpatients, who then take the drugs under observation. Such assurance of a complete TB treatment greatly reduces the rate of relapse, slows the evolution of resistance, and decreases the cost of treating resistant TB. It has been used in developing countries as well as in urban centers in the United States to help stem TB epidemics. The basic principle applies to any infectious disease for which a partial antibiotic dose would generate a strong evolutionary response to selection. The strategy slows arms races because it breaks the evolutionary cycle.
Integrated Pest Management
Attempts to slow the evolution of resistance are not restricted to medicine. Insect resistance to pesticides is a growing problem (Georghiou, 1986) in which arms races generate escalation in the strength and concentrations of insecticides used by farmers. Control failures result in increased pesticide use, potential environmental damage, and expensive crop losses. Reducing the speed of evolution in these cases can result from reducing the selection imposed by insecticides, a reduction that can be accomplished by using alternative tactics—besides chemical treatment—to control populations. Alternatives like using traps, baits, or an insect predator such as a praying mantis, or using crop rotations to prevent buildup of particular pest species can all postpone the need for chemical spraying, and therefore reduce the selection imposed by pesticides, and thus reduce the evolution of resistance. In these cases, overall insect mortality might remain high. But because the mortality sources are variable, the response to selection caused by any single mortality agent is smaller, and the evolution that results is slower.
In hospitals, integrated pest management can be called hand washing—the use of physical antimicrobial measures to reduce populations of potential infectious agents. These approaches do not supplant the use of antibiotics, but they reduce their use, saving them for times and places in which antibiotic drugs are absolutely required. Because antibiotic use is lower, selection for resistance is lower. Consider the result of using spray antibiotics to sterilize hospital instruments or other equipment. Each application acts as an opportunity for selection of bacterial populations for increased resistance. But when those same instruments are comprehensively sterilized with heat, there is no selection for antibiotic resistance.
Alternating Antibiotic Use
The general principle of diversifying the force of selection can apply to antimicrobial use if a suite of different agents are employed so that the selection against any single one of them is lessened. Like the above example in which antimicrobial use is lessened through application of other disinfectant techniques, selection for resistance to any one antimicrobial can sometimes be lessened through alternating use of different drugs. Both in medicine and agriculture, cyclic use of chemical control agents lessens the evolution of resistance. For herbicides, for example, many farmers are discouraged from using the same herbicide more than two growing seasons in a row, because otherwise resistant weeds crop up too often.
Likewise, some hospitals have tried cycling among different antibiotics in an attempt to reduce the evolution of resistance, and reduce the costs of treating resistant infections. In these cases, drug use is switched on a prescribed schedule, often resulting in reduced infections, reduced incidence of resistance, or reduced costs (see Table 1-2). Emerging theory helps design treatment protocols that limit resistance (Bonhoeffer et al., 1997), although substantial questions about how to implement these strategies remain.
TABLE 1-2
Responses to Antibiotic Cycling in Hospitals Trying to Reduce Costs of Treating Resistant Infections.
Fitness Costs of Resistance
Trade-offs in adaptation are a consistent theme in evolutionary science—adaptation to favor one attribute (like flight in birds) may have a high cost (e.g., large mass allocation to flight muscles) or lead to parallel changes that may be deleterious in some cases (like hollow and brittle bones). In cases in which flight was no longer adaptive—like predator-free oceanic islands—these fitness costs of flight are reversed, and birds have evolved flightlessness (a feature that is not necessarily a good strategy after humans discover these islands, as the dodo and many flightless New Zealand moas found out).
In the case of antimicrobial resistance, the evolution of mechanisms to evade drug actions might be highly costly or generate parallel, deleterious change. Enzymes that degrade or pump antibiotics are expensive to produce. Ribosomes that are resistant to tetracycline may be slower to synthesize proteins. If this were generally true, then cessation of use of an antibiotic would result in the de-evolution of resistance because cells that did not have resistant abilities would be favored. In such cases, we could reverse the course of evolution by shelving the drug for a while.
Although antibiotic resistance mechanisms that impose a fitness cost are known, in general fitness costs of antibiotic resistance are fairly small. In some experiments, resistant and non-resistant bacterial strains have been grown together in the absence of antibiotics for thousands of generations without the resistantless strains taking over. Such cases generate problems because once established, these “cheap” resistance mechanisms are difficult to eradicate. Antibiotic cycling protocols detailed above will not necessarily work to reduce the evolution of these strategies, and other approaches like Integrated Pest Management will have similarly reduced effectiveness.
We cannot change the nature of resistant mutations, and therefore cannot engineer the evolution of mutations that happen to have fitness costs. However, one avenue to explore is the development of new antibiotics for which a resistance mechanism would pose a fitness cost. The choice to develop and deploy an antibiotic rests on many different features of its performance, but one feature that might be taken more heavily into account is the nature and cost of mutations that provide resistance. Given that resistance is inevitable, those antibiotics that impose a fitness cost on resistant strains might be most valuable in the long term—both from a public health and from an economic point of view.
Conclusions
Evolutionary arms races provide a valuable conceptual framework in which to understand the nature of the current crisis in antimicrobial resistance, and understand the evolutionary principles involved. Even if coevolutionary changes are inevitable as a result of antimicrobial arms races, there are strategies that can be used to slow down their advance, and perhaps curtail the evolution of resistance. Current examples of successful deceleration of evolution include drug overkill strategies, direct observation therapy, integrated pest management, and antibiotic cycling. Use of these tactics follows from the realization that fighting infectious disease is only part of the health care battle. In addition, fighting the evolution of these diseases must be part of the agenda for long-term human health care.
ANTIBIOTIC RESISTANCE 1992–2002: A DECADE'S JOURNEY
Authors
Stuart B. Levy, M.D.1.Affiliations
This overview briefly examines the status of antibiotic resistance over a 10-year perspective which is described more fully in the second edition of The Antibiotic Paradox (Levy, 2002a).
In 1992, I warned about ominous events, like the emergence of vancomycin-resistant Staphylococcus aureus or the widespread appearance of Streptococcus pneumoniae resistant to multiple drugs, or fluoroquinolone-resistant E. coli (Levy, 1992). It is no surprise now that these are actual events—but what else has happened over the past decade?
While there are many factors involved in the emergence of antibiotic resistance, the two major ones still operate today: the antibiotic, which encourages selection and evolution of resistant strains, and the resistance genes which are acquired by bacteria or appear by mutation.
Over the past ten years, investigators have identified many additional transferable resistance genes. In 1992 there were 25 TEM-like β-lactamases described. Currently, the β-lactamase website (http://www.lahey.org/studies/webt.stm) lists over 100. In 1992, tetracycline resistance determinants numbered 12; in 2002, there are 35. Ten years ago, there were perhaps two or three types of multi-drug efflux pumps. Now, there are five (Alekshun and Levy, 2000). These new genetic findings represent growing knowledge of the breadth and complexity of antibiotic resistance.
These findings indicate that new drug resistance determinants are reaching clinically detectable levels under the selection of continued antibiotic misuse. Scientists are stimulated to evaluate, clone, and genetically define them. These efforts represent an increased interest in resistance—not nearly enough, but more than before. It is heartening to see laboratories defining new resistance genes and investigating their biochemical activity.
Over the past decade, the use of antibiotics has continued to increase, even though the actual quantity is still controversial. By my estimate there are between 35 and 50 million pounds of antibiotics being produced in the United States (Levy, 2002a). Some groups have said that animals use half this amount. Others say that almost 25 million pounds go to animals and that people use less than 5 million pounds (Mellon et al., 2001). Studies are underway to try to quantitate antibiotic use. In that context, an estimated 260 million prescriptions for antibiotics were filled in outpatient pharmacies in the United States in 1996 (Hutchinson, 1998). This figure does not include hospital use. Therefore, a considerable amount of pressure is being exerted on the natural microbial environment by the antibiotics provided to humans, animals, and plants, including the spraying of antibiotics on fruit trees.
One area in which antibiotic usage has decreased is in aquaculture. In 1992 there were no FDA regulations on how antibiotics were to be provided to the fish industry (Levy, 1992). There was no government mark on a piece of fish that identified whether it did or did not come from a fishery using antibiotics. Today the aquaculture industry has decreased the use of antibiotic-impregnated feeds by as much as four-fold (Levy, 2002a). Part of the reason for this change is in response to outside pressure; part is linked to improved hygiene and increased use of vaccines. The industry uses public areas to raise the fish. The seas, ponds, and other waters are overseen by ecology groups which have little tolerance for environmental pollution and put forth great efforts in protecting the environment.
An area that has also broadened over the decade is the genetics of resistance. We have heard about transposons, plasmids, and bacteriophages before, but a newer genetic entity has reached greater recognition in the last decade—the “integron.” Showing structural resemblance to transposons, integrons carry different genes, including those for antibiotic resistance, in a series behind a single promoter from which expression of all the genes downstream occurs. Integrons capture and integrate cassettes of antibiotic resistance genes and other genes via an integrase. The genes are joined in tandem, producing a single element mediating resistance to multiple antibiotics. The integron has been shown to be responsible for multi-drug resistance in a large number of gram-negative bacteria and helps explain the multi-drug resistance phenomenon of the current and previous decade.
At the end of the 1980s and continuing into the 1990s, vancomycin-resistant enterococci (VRE) bearing a large plasmid-borne transposon began to appear in hospital patients. The situation has worsened over the past decade. Virtually every hospital in the United States encounters VRE. It was expected that this genetic element would enter S. aureus, largely because transfer had been shown on mouse skins in the laboratory of W. Noble in London (Noble et al., 1992). But it did not happen quickly over the ensuing years, and, actually, not until just this year (2002).
Two different strains of S. aureus with full-fledged vancomycin resistance mediated by the vanA gene were isolated in the United States. The first was identified in a patient in Michigan in July (Centers for Disease Control and Prevention [CDC], 2002a) and the second in a patient in Pennsylvania in October (CDC, 2002b). Thus the drastic event feared a decade ago is now a reality. We can wonder what the future will bring in terms of other vanA-resistant S. aureus clinical isolates and how wide the determinant has already spread among staphylococci.
Before this current emergence, hospitals saw S. aureus strains that were heterogeneously resistant or homogeneously resistant to vancomycin by chromosomal mutation. These so-called vancomycin intermediate resistant strains, or “VISA,” have appeared in Japan, the United States, and elsewhere. Although the clinical laboratory tests (e.g., MIC values) might not designate these strains as fully resistant, patients bearing them have failed vancomycin therapy. Thus, they are clinically vancomycin-resistant.
In the beginning of the last decade, the fluoroquinolone family of antibiotics was a small group. Today there are more than a dozen different members of this family used worldwide. Initially, these agents were provided for serious multi-drug-resistant infections. Eventually, as resistance emerged to other first-line antibiotics, (for instance, in E. coli urinary tract infections), the fluoroquinolones became much more widely used in the community. Following closely on the heels of this increased use was the emergence of fluoroquinolone resistance. There are strains of E. coli in Southeast Asia, China, and even in the United States, that are resistant to greater than 250 μg/ml of the drug—the result of multiple mutations in the target genes, the topoisomerases, in combination with increased expression of multi-drug efflux pumps. What happened is something few could have predicted—the emergence of many different mutations producing a clinically relevant fluoroquinolone resistance in gram-negative bacteria.
Among the staphylococci, fluoroquinolone resistance was detected relatively soon after the introduction of the first fluoroquinolones. Gram-positive bacteria, including S. aureus, emerged with target gene mutations and/or efflux pumps. These mutations when present in a strain make the newer generation fluoroquinolones (designed to treat gram-positive bacteria) less effective and render the strains one or two mutational steps from full-fledged resistance.
In London, ciprofloxacin resistance in Salmonella typhi rose from just a few percent in 1996–1997 to about 25 percent in 1998 and 1999 (Threlfall and Ward, 2001). Many of the strains initially came from outside the United Kingdom, but were propagated and spread by fluoroquinolone use within the United Kingdom. This finding raises grave concerns that ciprofloxacin may not remain effective as a drug of last resort in those parts of the world where resistance to other antibiotics, namely ampicillin, chloramphenicol, and co-trimoxazole, has eliminated these drugs as effective treatment for typhoid fever.
In the United States, the CDC described four patients in Minnesota and North Dakota who died from a methicillin-resistant S. aureus (MRSA) infection whose resistance was not recognized early (CDC, 1999). MRSA, while well known among hospital patients, is not commonly found in the community. The unique feature of the so-called “community-acquired MRSA” was that they were resistant chiefly to β-lactam antibiotics, which included methicillin, but not other antibiotic classes like vancomycin, or the aminoglycosides.
For the physicians in the community, the first thought was to use a penicillin. No one expected that MRSA was the infectious cause in these children. The number of community-acquired MRSA strains is increasing: in one Chicago hospital, the rate per 100,000 admissions was 10 in 1988–1990, rising to 259 in 1993–1995 (Herold et al., 1998).
Also in the community, there has been a steady rise in antibiotic-resistant pneumococci. Just recently, the CDC reported an 11-month-old girl from a community in Georgia with refractory otitis media caused by a pneumococcus resistant to six different antibiotics (CDC, 2002c). She required hospitalization. The infection was traced to a day-care center where many other children were colonized with the same organism. This was not an urban environment with heavy antibiotic usage, but rather a small community. However, antibiotic usage there was largely without guidelines. Importantly, the pneumococcal vaccine, which covered the organism involved, had not been introduced.
Today, many different multi-drug-resistant bacteria, including both gram-negative and gram-positive bacteria, cause problems in communities as well as hospitals (see Table 1-3). These resistant organisms affect the health of our larger populations.
TABLE 1-3
Clinical Problems of Multi-Drug Resistance.
What else has happened to encourage resistance?
Antibacterials is a common name given to surface agents (otherwise known as biocides) contained within household products which inhibit a variety of microbes. Some act quickly like alcohols, peroxides, and chlorinated products. Others, such as triclosan or benzalkonium chloride, leave residues. The latter encourage the selection of resistant organisms. In 1992 there were 23 such products on the market; in 1999, The New York Times estimated 700 of these items marketed for households. The public is amassing these antibacterials in their homes in a variety of products. Why? To some extent it is in response to fear of the “super bug,” the multi-drug-resistant bacterium. The other pressure is to protect the family against infectious organisms caused by food. “I want to do what is best for my child,” is commonly expressed. While these reasons cannot be criticized, the public needs to be informed that the same protection is provided by using standard soap and disinfectants.
The antibacterial craze demonstrates a couple of problems. For one, it defies the message that washing with soap and water is sufficient to provide hygiene to healthy individuals. Today's public believes one has to add an antibacterial chemical to gain a health benefit from washing. This opinion does not help our cause against antibiotic misuse and the emergence of resistance.
Secondly, bacteria emerging with resistance to these chemicals can show decreased susceptibility to antibiotics. A Japanese group reported a few years ago that MRSA selected in the laboratory for half the susceptibility to a biocide, benzalkonium chloride, showed a dramatic increase in resistance to a wide spectrum of β-lactam antibiotics (Akimitsu et al., 1999). Susceptibility to other antibiotics was either unchanged or changed minimally. This phenotype, that is, predominantly β-lactam resistance, is strikingly similar to the community-acquired MRSA strains. Is it possible that disinfectants, not β-lactam antibiotics, selected these resistant bacteria?
S. aureus is one of the principal organisms targeted by triclosan in the hospital. But decreased susceptibility to triclosan is emerging in MRSA isolated from hospitals (Suller and Russell, 2000). This evolution will curtail this important use, fail to protect from transmission of disease, and create more resistance problems. A mutation in the target gene of triclosan activity, fabI (enoyl reductase) affects other drugs besides triclosan (McMurry et al., 1998). Given the multi-drug efflux pumps in E. coli and S. aureus, overexpression of one of these pumps, like in E. coli and Pseudomonas (Chuanchuen et al., 2001), could contribute to resistance mediated by a fabI mutation. These pumps export antibiotics as well. So, one selects a resistance mechanism affecting both antibacterials and antibiotics.
What does the future look like? What is happening today that provides optimism for the future?
When we entered the 1990s, the only in-depth analysis of antibiotic use and resistance was a report from an NIH Fogarty International Center-supported Task Force effort in 1983–1986 (Levy et al., 1987). Over 100 physicians, scientists, and public health and industry representatives from all over the world discussed the imminent problems of resistance through six task forces. Although the report urged that something be done about the problem, the effort was disregarded and even downplayed under the guise that the problem was being overstated. Some influential critics from industry believed that such an activity cast wrongful doubts on the efficacy of their antibiotic products. The report was not accepted as hoped—as a call to action by physicians, consumers, and public health officials.
More recently in 1995, the U.S. Office of Technology Assessment (OTA) released an assessment of the resistance problem in the United States just before the Office was closed (OTA, 1995). It stated the obvious—resistance was a mounting public health problem stemming from antibiotic misuse. The report particularly was noteworthy because Congress was showing interest at last. Since then, there have been reports from other professional and government groups including the American Society for Microbiology (ASM, 1995), the Society for Hospital Epidemiology of America (Shlaes et al., 1997), the American College of Physicians-American Society of Internal Medicine (ACP-ASIM, 2001), and the House of Lords Select Committee on Science and Technology (1998). We have learned something from these efforts, but up until now, they have had little impact on the problem.
Somewhat greater success has been achieved by The Alliance for Prudent Use of Antibiotics (APUA), founded in 1981, working in conjunction with its 35 country chapters on research and education concerning antibiotic use and resistance. For 20 years, it has championed the improved use of antibiotics globally. Recently, the organization synthesized the efforts of various expert groups—25 reports over the past 15 years—as a background and foundation for WHO's Global Strategy for Containment of Antimicrobial Resistance (WHO, 2001). The APUA document was published along with the WHO report in 2001 (APUA, 2001).
APUA has created materials to educate the consumer about antibiotics and resistance. Its “antibiotic” brochure has been translated into several languages and has been disseminated to almost a million people. The newsletter, translated into Russian and Spanish, reaches thousands quarterly. The CDC has also published a pamphlet for patients, which appeals to the consumer to stop demanding and stockpiling antibiotics.
The most important thing that has happened in this past decade is the wider awareness and response to the resistance problem. The WHO General Assembly selected resistance as one of the three major health problems facing the world today. The ACP-ASIM has chosen antibiotic resistance as its clinical theme for 2000–2002.
Are we going to see a difference? I hope so, but forums like this one have to define practical ways in which change can occur. We do not need more reports; we need action.
The pharmaceutical companies left the antibiotic field in the mid-1980s, came back to it in the mid-1990s, but appear to be leaving it again now. Antibiotics are difficult and costly to develop. Sales may not reach the minimum of $500 million to $1 billion per year to maintain the interest of the large pharmaceutical companies. This is a sad situation, complicated by the fact that many large companies have merged to form even larger ones. Thus, the creativity and competition that occurred among companies before is greatly diminished as, for instance, three companies become one. It is more likely now that new antibiotics will come from the smaller biopharmaceutical groups that make firm commitments to the discovery efforts. Big pharmaceutical companies join when the drug is far enough along that the risk is minimized.
Vaccination has made another big impact over the last decade. The involvement of Haemophilus influenzae type B in otitis media and meningitis has been curtailed with the use of the conjugate vaccine. We must expand acceptance of vaccination with the pneumococcal conjugate. A recent report looked at the effects of the vaccine coverage on invasive disease and found the vaccine to be very effective (Black et al., 2001). While less than 20 percent were fully vaccinated, a much larger percent of the individuals had reduction in invasive disease indicating that there was a “herd effect.” If we can get the vaccine to more communities, patients like that little 11-month-old girl from Georgia can be protected. We would greatly reduce pneumonia and prevent fatal meningitis. We also would decrease the initial choice of an antibiotic for a suspected bacterial etiology (e.g., pneumococcus) of upper respiratory disease like otitis media. Providing vaccines against both H. influenzae and S. pneumoniae would make a viral cause of the illness more acceptable and antibiotics less prescribed.
Another initiative at APUA is the Global Advisory on Antibiotic Resistance Data (GAARD), which includes the large surveillance systems of Bristol-Myers Squibb (SENTRY), GlaxoSmithKline (ALEXANDER), Focus Technology, and, most recently, AstraZeneca (MYSTIC) and Bayer. Large amounts of data on drug-bug combinations are collected which can be helpful to public health officials. APUA was able to catalyze an agreement between the companies funding these projects to meet periodically and discuss and share the data. The project provides antibiotic susceptibility surveillance information on large numbers of clinical isolates and allows early changes in susceptibility to be revealed, such as the decreased susceptibility of Haemophilus influenzae to fluoroquinolones detected at 0.1 percent (Travers et al., 2001).
Another project that has recently been completed, also by APUA, is an in-depth examination of the impact of animal use of antibiotics on human public health and the environment (APUA, 2002). The FAAIR (Facts About Antibiotics in Animals and the Impact on Resistance) project brings scientific evidence to the policy debate regarding use of antibiotics for animals. A unique feature of this report is its broader perspective. It looks at the ecological impact of animal use, not just the direct animal-to-consumer path of transmission. Antibiotics provided to animals for therapy, prophylaxis, or as growth promotion, select resistant organisms which can spread to farm dwellers, as well as food consumers.
The important, but often lost, message is that antibiotics affect more bacteria than the targeted pathogen. They will decimate entire microbial communities that are exposed. Those that are resistant become reservoirs of antibiotic resistance genes.
In fact, an antibiotic provided to humans, plants, or animals does not stay with the treated individual nor is it always readily degraded. It eventually passes into the environment in some form at some level, which explains the detection of drugs in municipal waters, streams, and agricultural fields (Kolpin et al., 2002). We can now speak not only of antibiotic effects during therapy, but effects “after therapy” (Levy, 2001, 2002a). Since resistance does not often emerge in the person while on the antibiotic, I suggest that much resistance occurs outside the treated person or animal. Perhaps it would be considered unrealistic, but what an advantage there would be to develop an antibiotic that does its treatment, but then self-destructs.
With this point in mind and focusing on the environment, APUA and University of Illinois, funded by the National Institute of Allergy and Infectious Diseases, began the Reservoirs of Antibiotic Resistance (ROAR) project. The aim is to look for the genes of resistance harbored in natural environments as they may evolve as future clinical treatment problems.
Can we reverse the resistance problem? It may take a long time for a bacterium to lose a transposon or a mutation, but one way to reverse resistance is to encourage the susceptible commensal organisms to return. Such was the basis for reversal of macrolide resistance among Streptococcus pyogenes in Finland when physicians stopped the use of erythromycin (Seppala et al., 1997). It was not due to loss of resistance from a particular S. pyogenes, but, rather, by replacement with other susceptible strains.
In addition to the various scientific challenges that must be met, we also need to devise new public policies and procedures that will help minimize the emergence of antimicrobial resistance. For example, I have been suggesting for a number of years that the Food and Drug Administration (FDA) create an entirely separate category for antibiotics, apart from other drugs. Antibiotics are the only type of therapeutic agents that have societal effects beyond their physiological effects, so it is logical for them to have their own official category, complete with its own set of regulatory requirements. The FDA would then be better positioned to devise unique incentives that encourage the pharmaceutical industry to pursue the development of new antibiotics, to market antibiotics prudently, and to conduct surveillance efforts to see that any incipient antimicrobial resistance is contained. As we use antibiotics in this decade, we should be thinking more ecologically. Let our treatments be directed at the causative agents and less destructive of the broad microbial environment.
REFERENCES
- Abramson MA, Sexton DJ. Nosocomial methicillin-resistant and methicillin-susceptible Staphylococcus aureus primary bacteremia: at what cost? Infection Control and Hospital Epidemiology. 1999;20:408–411. [PubMed: 10395142]
- ACP-ASIM (American College of Physicians-American Society of Internal Medicine). Principles of Appropriate Antibiotic Use Position Papers. Annals of Internal Medicine. 2001;134:479–529. [PubMed: 11255524]
- Akimitsu N, Hamamoto H, Inoue R, Shoji M, Akamine A, Takemori K, Hamasaki N, Sekimizu K. Increase in resistance of methicillin-resistant Staphylococcus aureus to beta-lactams caused by mutations conferring resistance to benzalkonium chloride, a disinfectant widely used in hospitals. Antimicrobial Agents and Chemotherapy. 1999;43:3042–3043. [PMC free article: PMC89614] [PubMed: 10651623]
- Alekshun MN, Levy SB. Bacterial drug resistance: response to survival threats. In: Storz G, Heugge-Aronis R, editors. Bacterial Stress Responses. Washington, DC: ASM Press; 2000. pp. 323–366.
- APUA (Alliance for the Prudent Use of Antibiotics). Antibiotic Resistance: Synthesis of Recommendations by Expert Policy Groups. Geneva: WHO; 2001. WHO/CDS/CSR/DRS/2001.10.
- APUA. The need to improve antimicrobial use in agriculture: ecological and human health consequences. Clinical Infectious Diseases. 2002;34(Suppl 3):S71–S144.
- Ash RJ, Mauck B, Morgan M. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerging Infectious Diseases. 2002;8:713–716. [PMC free article: PMC2730334] [PubMed: 12095440]
- ASM (American Society for Microbiology). Report of the ASM task force on antibiotic resistance. Antimicrobial Agents and Chemotherapy. 1995;(Suppl):S1–S23. [PubMed: 7654082]
- Black SB, Shinefield HR, Hansen J, Elvin L, Laufer D, Malinoski F. Postlicensure evaluation of the effectiveness of seven valent pneumococcal conjugate vaccine. Pediatric Infectious Diseases Journal. 2001;20:1105–1107. [PubMed: 11740313]
- Bonhoeffer S, Lipsitch M, Levin B. Evaluating treatment protocols to prevent antibiotic resistance. Proceedings of the National Academy of Sciences. 1997;94:12106–12111. [PMC free article: PMC23718] [PubMed: 9342370]
- Bozzette SA, Joyce G, McCaffrey D, Leibowitz A, Morton S, Berry S, Rastegar A, Timberlake D, Shapiro M, Goldman D. Expenditures for the care of HIV-infected patients in the era of highly active antiretroviral therapy. New England Journal of Medicine. 2001;344:817–823. [PubMed: 11248159]
- CDC (Centers for Disease Control and Prevention). Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. Morbidity and Mortality Weekly Report. 1999;48:707–710. [PubMed: 21033181]
- CDC. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morbidity and Mortality Weekly Report. 2002;51:565–567. [PubMed: 12139181]
- CDC. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morbidity and Mortality Weekly Report. 2002;51:902. [PubMed: 12418544]
- CDC. Multidrug-resistant Streptococcus pneumoniae in a child care center—Southwest Georgia, December 2000. Morbidity and Mortality Weekly Report. 2002;50:1156–1158.
- Chuanchuen R, Beinlich K, Hoang TT, Karkhoff-Schweizer RR, Schweizer HP. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrobial Agents and Chemotherapy. 2001;45:428–432. [PMC free article: PMC90308] [PubMed: 11158736]
- Consumers Union of the United States. Super-germ alert: how to avoid antibiotic use and misuse. Consumer Reports. 2001;66:60–61. [PubMed: 11503747]
- Crandall K, editor. The Evolution of HIV. Baltimore: Johns Hopkins University Press; 1999.
- Dominguez EA, Smith TL, Reed E, Sanders CC, Sanders WE Jr. A pilot study of antibiotic cycling in a hematology-oncology unit. Infection Control and Hospital Epidemiology. 2000;21(1 Suppl):S4–8. [PubMed: 10654628]
- Garrett L. The Coming Plague: Newly Emerging Diseases in a World out of Balance. New York: Farrar, Straus and Giroux; 1994.
- Georghiou GP. Pesticide Resistance: Strategies and Tactics for Management. Washington, DC: National Academy Press; 1986. The magnitude of the resistance problem; pp. 14–43.
- Gerding DN, Larson TA, Hughes RA, Weiler M, Shanholtzer C, Peterson LR. Aminoglycoside resistance and aminoglycoside usage: ten years of experience in one hospital. Antimicrobial Agents and Chemotherapy. 1991;35:1284–1290. [PMC free article: PMC245159] [PubMed: 1929283]
- Gruson D, Hilbert G, Vargas F, Valentino R, Bebear C, Allery A, Bebear C, Gbikpi-Benissan G, Cardinaud JP. Rotation and restricted use of antibiotics in a medical intensive care unit. Impact on the incidence of ventilator-associated pneumonia caused by antibiotic-resistant gram-negative bacteria. American Journal of Respiratory and Critical Care Medicine. 2000;162:837–843. [PubMed: 10988092]
- Hastings IM, Mackinnon MJ. The emergence of drug-resistant malaria. Parasitology. 1998;117:411–417. [PubMed: 9836305]
- Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. Journal of the American Medical Association. 1998;279:593–598. [PubMed: 9486753]
- Ho D. Time to hit HIV, early and hard. New England Journal of Medicine. 1995;333:150–151. [PubMed: 7616996]
- Honore N, Cole ST. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrobial Agents and Chemotherapy. 1993;337:414–418. [PMC free article: PMC187686] [PubMed: 8460911]
- House of Lords Select Committee on Science and Technology. Seventh Report. 1998. [Online]. Available: http://www
.parliament .the-stationery-office .co.uk/pa/ld199798 /ldselect/ldsctech/081vii/st0701.htm. - Hutchinson JM. More than 160,000,000 antibiotic prescriptions in the USA. Abstract O-22. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy; San Diego. September 24–27, 1998.
- Kollef MH, Vlasnik J, Sharpless L, Pasque C, Murphy D, Fraser V. Scheduled change of antibiotic classes: a strategy to decrease the incidence of ventilator-associated pneumonia. American Journal of Respiratory and Critical Care Medicine. 1997;156:1040–1048. [PubMed: 9351601]
- Kollef MH, Ward S, Sherman G, Prentice D, Schaiff R, Huey W, Fraser VJ. Inadequate treatment of nosocomial infections is associated with certain empiric antibiotic choices. Critical Care Medicine. 2000;28:3456–3464. [PubMed: 11057801]
- Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environmental Science and Technology. 2002;36:1202–1211. [PubMed: 11944670]
- Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science. 1989;246:1155–1158. [PubMed: 2479983]
- Levy SB. The Antibiotic Paradox: How Miracle Drugs Are Destroying the Miracle. New York: Plenum Publishing; 1992.
- Levy SB. Antibiotic resistance: consequences of inaction. Clinical Infectious Diseases. 2001;33:S124–S129. [PubMed: 11524708]
- Levy SB. The Antibiotic Paradox: How the Misuse of Antibiotics Destroys Their Curative Powers. Boston: Perseus Books; 2002.
- Levy SB. The 2000 Garrod lecture: factors impacting on the antibiotic resistance process. The Journal of Antimicrobial Chemotherapy. 2002;49:25–30. [PubMed: 11751763]
- Levy SB, Burke JP, Wallace CK. Antibiotic use and antibiotic resistance worldwide. Report of a study sponsored by the Fogarty International Center of the National Institutes of Health, 1983–1986. Reviews of Infectious Diseases. 1987;9 Supplement 3:S231–316. [PubMed: 3602801]
- Lewis R. The rise of antibiotic-resistant infections. FDA Consumer. 1995;29:11–15. [PubMed: 10145087]
- McMurry LM, Oethinger M, Levy SB. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. [PubMed: 9707111]
- Mellon M, Benbrook C, Benbrook KL. Hogging It. Cambridge, MA: Union of Concerned Scientists; 2001.
- Neu HC. Emerging trends in antimicrobial resistance in surgical infection: a review. European Journal of Surgery Supplement. 1994;(573):7–18. [PubMed: 7524800]
- Noble WC, Virani Z, Cree RG. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiology Letters. 1992;72:195–198. [PubMed: 1505742]
- OTA (Office of Technology Assessment). Impacts of Antibiotic-Resistant Bacteria. Washington, DC: OTA; 1995.
- Palumbi SR. The Evolution Explosion: How Humans Cause Rapid Evolutionary Change. New York: WW Norton; 2001.
- Palumbi SR. Humans as the world's greatest evolutionary force. Science. 2001;293:1786–1790. [PubMed: 11546863]
- Rubin RJ, Harrington C, Poon A, Dietrich K, Greene J, Moidussin A. The economic impact of Staphylococcus aureus infection in New York City hospitals. Emerging Infectious Diseases. 1999;5:9–18. [PMC free article: PMC2627695] [PubMed: 10081667]
- Schentag JJ, Hyatt JM, Fitzpatrick P, Paladino JA, Birmingham MC. Infection control and changes in the antibiotic formulary for management of epidemic and endemic vancomycin-resistant Enterococcus faecium. Hospital Practice 1998 Special Report. 1998:22–36.
- Seppala H, Klaukka T, Vuopio-Varkila J, Muotiala A, Helenius H, Lager K, Huovinen P. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. Finnish Study Group for Antimicrobial Resistance. New England Journal of Medicine. 1997;337:441–446. [PubMed: 9250845]
- Shlaes DM, Gerding DN, John JF, Craig WA, Bornstein DL, Duncan RA, Eckman MR, Farrer WE, Greene WH, Lorian V, Levy S, McGowan JE Jr, Paul SM, Ruskin J, Tenover FC, Watanakunakorn C. Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the Prevention of Antimicrobial Resistance: Guidelines for the Prevention of Antimicrobial Resistance in Hospitals. Infection Control and Hospital Epidemiology. 1997;18:275–291. [PubMed: 9131374]
- Suller MT, Russell AD. Triclosan and antibiotic resistance in Staphylococcus aureus. Journal of Antimicrobial Chemotherapy. 2000;46:11–18. [PubMed: 10882683]
- Threlfall EJ, Ward LR. Decreased susceptibility to ciprofloxacin in Salmonella enterica serotype Typhi, United Kingdom. Emerging Infectious Diseases. 2001;7:448–450. [PMC free article: PMC2631792] [PubMed: 11384525]
- Travers K, Stelling J, Levy SB, Miller L, Souder B, Jones R. Emerging geographically diverse Haemophilus influenzae isolates with reduced susceptibility to fluoroquinolones: a GAARD report. Late breaker session at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy; Chicago. December 16–19, 2001.
- WHO (World Health Organization). Global Strategy for the Containment of Antimicrobial Resistance. Geneva: WHO; 2001. WHO/CDS/CSR/DRS/2001.2.
- Wolinsky SM, Korber BTM, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT, Koup RA. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–541. [PubMed: 8614801]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. [PubMed: 9360926]
- Introduction - The Resistance Phenomenon in Microbes and Infectious Disease Vect...Introduction - The Resistance Phenomenon in Microbes and Infectious Disease Vectors
Your browsing activity is empty.
Activity recording is turned off.
See more...