The content of this book is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 Unported license. To view the terms and conditions of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology [Internet]. 4th edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. doi: 10.1101/glycobiology.4e.23
Fungi are a fascinating group of predominantly multicellular organisms. Fungal species, such as Saccharomyces cerevisiae, have been instrumental in defining the fundamental processes of glycosylation, but their glycobiology is significantly different from animal or plant systems. This chapter describes the glycan structures that compose the fungal cell wall, offers some insights into novel glycobiology revealed through studying fungal systems, addresses the use of fungi as experimental and synthetic systems, and delineates the relationships of several important glycoconjugates to fungal biology and pathogenesis.
FUNGAL DIVERSITY
More than 70,000 species of fungi have been described, and it is estimated that more than 5,000,000 fungal species exist. The fungal phyla are the Chytridiomycota (zoosporic fungi), the Opisthosporidia, the Neocallimastigomycota, the Blastocladiomycota, the Zoopagomycota, the Mucoromycota, the Glomerulomycota (abuscular mycorrhizal fungi), the Ascomycota (sac fungi, e.g., Saccharomyces, Candida, Aspergillus, Neurospora, and morel mushrooms), and the Basidiomycota (e.g., mushrooms, rot fungi, and puffballs). The vast majority of species belong to the Ascomycota phylum, which together with the Basidiomycota form the subkingdom Dikarya and comprise the most studied species; this chapter focuses on model organisms in these two clades. Most fungi are primarily made of hyphae (branching filaments) that form the mycelium and multicellular structures such as fruiting bodies, whereas the alternative fungal life-form is growth as unicellular yeast. The extracellular matrix of all fungi, the cell wall, comprises complex polysaccharides including mannans, galactans, glucans, and chitin and represents a major target of fungicides.
FUNGI AS MODEL SYSTEMS FOR GENETICS, BIOCHEMISTRY, AND GLYCOBIOLOGY
Historical Perspective
More than 100 years ago, Louis Pasteur discovered that fermentation requires a viable organism; since then yeast have been used as a model system to study cellular metabolism. In fact, Pasteur coined the word “ferment” during his work on alcohol production by S. cerevisiae, or baker's yeast. This organism has been a tremendous resource for biologists and glycobiologists, especially because many of the fundamental enzymes in aerobic and anaerobic metabolism (terms also invented by Pasteur) are shared between yeast and animals. Breakthroughs in enzymology occurred following the 1897 discovery by the Buchner brothers that extracts of yeast could make ethanol and carbon dioxide from glucose, just like intact cells. Mannose is a major component of the yeast cell wall; it was discovered by Emil Fischer in 1888, and the mannose-rich glycans in yeast, historically called yeast gum, have been known since the 1890s. The discovery that the yeast cell wall was composed of D-mannose and work elucidating the chemical structures of other carbohydrates (and vitamin C) led to Sir Walter Norman Haworth's 1937 Nobel Prize in Chemistry. Luis Leloir subsequently discovered the activated precursors required for carbohydrate synthesis, identifying UDP-glucose, GDP-mannose, and other nucleotide sugars from yeast extracts. He was awarded the 1970 Nobel Prize in Chemistry for this work. The discovery of heterothallic yeast strains and the subsequent development of the field of yeast genetics led to multiple ground-breaking discoveries. For example, genetic studies initiated by the laboratory of Phil Robbins led to the molecular characterization of the conserved N- and O-glycosylation pathway in the endoplasmic reticulum (ER) and the biosynthesis of glycosylphosphatidylinositol (GPI)-anchored proteins. Yeast secretory (sec) mutants helped define the protein secretory pathway, by which polypeptides travel from the ER through the Golgi apparatus to the cell surface or surrounding milieu, becoming glycosylated en route. This foundational work in cell biology was recognized by the Nobel Prize in Physiology or Medicine awarded to Randy Schekman in 2013.
The Fungal Cell Wall
The fungal cell wall, like the plant cell wall, is composed of highly cross-linked glycan polymers (Figure 23.1), which adapt to growth conditions in a dynamic and flexible way and provide high mechanical stability. In contrast to the plant cell wall, it is directly connected to the plasma membrane and specific cell-wall polysaccharides of fungi differ from those of plants. Fungal cell walls are composed of glycoproteins and complex polysaccharides such as chitin, glucans, mannans, galactomannans, glucomannans, rhamnomannans, and phosphomannans. The nature and relative abundance of cell wall polymers varies between fungal species.
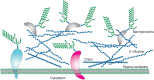
FIGURE 23.1.
Illustration of the cell wall of yeasts, showing glycan polymers and mannoproteins. The presence and abundance of different glucans and chitin varies between different fungal species.
Chitin is a polymer of β1-4-linked GlcNAc, which occurs in chains that typically exceed 1000 residues. These chains self-associate to form microfibrils and are deposited primarily at the bud neck of yeast or at septa in filamentous fungi. Chitin is produced by the regulated and coordinated action of multiple chitin synthases, which ensure the timely deposition at specific sites required for normal cell growth and division. Chitin may also be deacetylated to form the cationic polymer, chitosan.
β1-3 Glucan, synthesized from UDP-glucose at the plasma membrane, is a major polysaccharide of fungal cell walls, where it is cross-linked to chitin. A variety of glucans with other linkages, including β1-6, mixed β1-3/β1-4, α1-3, and α1-4, also occur in fungi. β1-6 glucan is a major cell wall component of the yeasts S. cerevisiae, Candida albicans, and Cryptococcus neoformans. In contrast, the filamentous fungi Aspergillus fumigatus and Neurospora crassa lack β1-6 glucans but synthesize a mixed β1-3/β1-4-glucan. α1-3 Glucan is also a common component of the cell wall of Ascomycota and some Basidiomycota, although it is absent from S. cerevisiae.
β-glucan chains act as attachment points for an external glycoprotein layer. The majority of these cell wall proteins are GPI-dependent and carry N- and O-linked glycans. In yeast cell wall proteins, the conserved N-glycan core structure is further elaborated with an extensive repeating α1-6-linked mannose chain. This chain is typically branched by short α1-2- and α1-3-linked mannose structures; some of these may be in phosphodiester linkage (Figure 23.2). These N-glycans are highly heterogeneous in length and branching. In contrast, filamentous fungi and Basidiomycota synthesize small oligomannose N-glycans, which may carry substituents such as galactofuranose, N-acetylglucosamine, xylose or fucose. In addition, fungal cell wall proteins bear Ser/Thr-linked O-mannose glycans (Figure 23.3A). During cell wall assembly, cell wall proteins are linked to the β-glucan via a GPI-remnant and/or via their N- and O-glycans. Cell wall construction is temporally and spatially controlled during the cell cycle, determining cellular shape (hyphae vs. yeast) and function.

FIGURE 23.2.
Structures of selected yeast mannans. Note that a single pyruvate is (R) 4,6 acetyl-(ketal)-linked to the terminal galactose residue in the pyruvylated structure.

FIGURE 23.3.
Structures of selected O-linked glycans in fungi: (A) yeast, (B) filamentous fungi, and (C) Cryptococcus.
Protein Glycosylation
Glycoproteins are a major component of the fungal cell wall, often bearing N- and O-glycans, as well as GPI anchors. As outlined in Chapter 9, N-glycan synthesis begins with synthesis of the conserved lipid-linked core glycan donor, Glc3Man9GlcNAc2-P-P-Dol, which is transferred to nascent polypeptides in the ER. Following core N-glycosylation, the Glc3Man9GlcNAc2Asn-R is processed with removal of glucose residues by α-glucosidases I and II to generate Man9GlcNAc2Asn-R. In mammals and S. cerevisiae, the Man9GlcNAc2Asn-R is further trimmed to Man8GlcNAc2-Asn-R by an ER-mannosidase. Schizosaccharomyces pombe, however, lacks this enzyme and stops processing at Man9GlcNAc2Asn-R. Man8GlcNAc2Asn-R in mammals and Man9GlcNAc2Asn-R in S. pombe are then substrates for the UDP-Glc: glycoprotein glucosyltransferase (UGT) that generates Glc1Man8GlcNAc2Asn and Glc1Man9GlcNAc2Asn in mammals and S. pombe, respectively. This reglucosylation, which is part of the quality control system for protein folding in the ER, is absent in S. cerevisiae (Chapter 39). The monoglucosylated structure is a ligand for the chaperone lectins calnexin and calreticulin in mammalian cells. Most fungi express calnexin but lack a calreticulin homolog. Specific trimming of the N-linked glycan regulates ER-associated degradation (ERAD) of improperly folded proteins or unassembled protein complex units. In S. cerevisiae, trimming of the N-linked glycan to Man7GlcNAc2 by the mannosidase Htm1p generates a signal that is recognized by the lectin Yos9p and leads to export of the glycoprotein to the cytoplasm and subsequent degradation. Notably, quality control and ERAD processes differ between fungal species; in some cases, components of the complete pathway are absent.
Yeast N-glycans are extended in the Golgi apparatus by mannosyltransferases that use GDP-Man as the donor, with one or more specific glycosyltransferases acting to catalyze synthesis of each linkage and branch.
Fungal proteins are rich in O-linked mannose. This protein modification is initiated by ER protein mannosyltransferases (PMTs) that use Dol-P-Man as a mannose donor. There are several hetero- or homodimeric PMTs in fungi and each may have a different substrate specificity and glycoprotein preference. The Dol-P-Man for the fungal PMTs is synthesized in the cytosol and then flipped into the secretory organelle lumen (Figure 23.4, top right) to be used for both N- and O-glycosylation. Subsequent additions of mannose residues to growing chains occur in the Golgi apparatus, where GDP-Man serves as the donor for reactions catalyzed by Mn++-dependent mannosyltransferases.
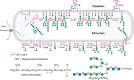
FIGURE 23.4.
Biosynthesis of N-glycans and their transfer to -Asn-X-Ser/Thr- sequons of newly synthesized glycoproteins in the fungal endoplasmic reticulum (ER). Individual steps in the biosynthesis pathway from dolichol phosphate (simplest structure at top right (more...)
Fungi express abundant GPI-anchored glycoproteins. As in other systems (Chapter 12), GPI-anchored proteins are synthesized in the ER from a GPI-precursor and a protein precursor having a carboxy-terminal GPI-addition signal peptide. A GPI-transamidase cleaves the signal peptide and replaces it by the GPI precursor. In fungi, addition of an α1-2 mannose residue to the conserved GPI core Manα1-2Manα1-6Manα1-4GlcNα1-6Inositolphospholipid by the enzyme Smp3 is a prerequisite for addition of the ethanolamine phosphate bridge that later carries the protein, although this step is dispensable in mammals. Consequently, fungal GPI-anchors have an extended core with four mannose residues (Figure 23.5). In an interesting divergence from “higher” eukaryotes, yeast GPI anchors may serve as substrates for transglycosylation reactions in cell wall assembly, leading to covalent linkage of glycoproteins to the glucan matrix of the cell wall. Polysaccharides linked to GPI anchors are also found in the extracellular matrix of filamentous ascomycetes.
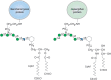
FIGURE 23.5.
Structures of two fungal glycosylphosphatidylinositol (GPI) anchors. Hexagon indicates myo-D-inositol.
Glycolipids
Yeast express a relatively simple array of glycolipids, although C. albicans is notable for its large lipid-linked mannans. Many fungi make short-chain glycolipids, commonly containing myo-inositol phosphate linked to mannose that may be modified by galactofuranose (as in Histoplasma capsulatum or A. fumigatus) or additional mannose residues. S. cerevisiae generates forms with a single residue of mannose, whereas some longer galactose- and mannose-containing glycolipids are found in Aspergilli. Short-chain glycosylceramides such as Glc-Cer and Gal-Cer are also found in the fungi Schizophyllum commune and A. fumigatus.
MODEL FUNGI
Saccharomyces cerevisiae as an Experimental System
Yeasts have been valued for baking and brewing for thousands of years, but in the last century or so scientific attention has particularly focused on S. cerevisiae, an oval budding yeast 5–10 µm across. The rapid growth of this simple eukaryote, combined with its inexpensive culture and genetic tractability, has made it a powerful and popular model system. Studies of S. cerevisiae have enormously influenced the fields of eukaryotic cell biology and genetics, in addition to their impact on basic metabolism and enzymology noted above.
S. cerevisiae contributed to defining the enzymology of GPI lipid precursor biosynthesis (Chapter 12). This complex process, involving more than 20 genes, presented a significant biochemical challenge to researchers in the field. However, as many of the steps are conserved from yeast to mammals, analysis of S. cerevisiae mutants offered a complementary and powerful approach to its dissection. Mutants have also been useful for dissecting yeast-specific processes such as mannan synthesis; this was elucidated by identifying mnn mutants, which displayed aberrant antibody or dye binding.
Despite the tremendous value of S. cerevisiae as a model, it does have certain limitations. These cells do not synthesize complex N-glycans, mucins or mucin-type O-glycans, O-linked N-acetylglucosamine (O-GlcNAc), sialic acids, or glycosaminoglycans (GAGs) of the types found in vertebrates. However, S. cerevisiae cells use O-mannose on their nucleocytoplasmic proteins in a manner analogous to O-GlcNAc in plants and animals. Like most other fungi, S. cerevisiae also lacks long-chain glycolipids (apart from those participating in the GPI synthesis) and does not synthesize complex glycosphingolipids or gangliosides like those found in mammals (although it is still valuable for studying sphingosine and sphingolipid metabolism). S. cerevisiae expresses limited glycan diversity even compared with other fungi, with no galactose, xylose, or glucuronic acid reported in its glycans. This must be kept in mind when generalizing from this model to other organisms.
Schizosaccharomyces pombe, a Model for Ultrastructure
S. pombe is a rod-shaped yeast, ∼3–4 µm in diameter and 7–14 µm long. Rather than budding, this organism grows by elongation and fission to yield equal-sized daughter cells. Like S. cerevisiae, it has a relatively small genome of ∼14 million base pairs. S. pombe has been useful as a genetically manipulatable model organism for studying the cell cycle. Because it has well-defined organelle structures compared with other yeast, it is a popular choice for studies of intracellular structure. S. pombe also synthesizes mannoproteins and mannans, some containing galactose (Figure 23.3A); this may occur as α1-2-linked caps that are also sometimes pyruvylated. The galactose residues are important for lectin recognition in nonsexual flocculation (clumping) of S. pombe, as evidenced by inhibition of this process by free galactose. In contrast, flocculation in S. cerevisiae is mannose-dependent and inhibited by free mannose. Another difference between S. pombe and the more common S. cerevisiae model is that its newly-synthesized N-glycans (Man9GlcNAc2Asn; Figure 23.4) are not trimmed to Man8GlcNAc2Asn in the ER.
HARNESSING YEAST FOR PRODUCTION
Pichia pastoris and Its Advantages for Expression
Pichia pastoris is a methylotrophic, nonpathogenic organism that was discovered in 1969 in a screen for yeast capable of using methanol. In this yeast, methanol is oxidized to formaldehyde and hydrogen peroxide by alcohol oxidase (AOX) in the peroxisome. The formaldehyde exits the peroxisome and is oxidized to formate and carbon dioxide in the cytoplasm for energy production. Any remaining formaldehyde is assimilated into glyceraldehyde-3-phosphate and dihydroxyacetone by condensation with xylulose-5-monophosphate, in a reaction catalyzed by the peroxisomal enzyme dihydroxyacetone synthase. P. pastoris has become popular as a model system for making recombinant proteins because it is easy to manipulate genetically and can be grown to very high densities. The promoter for AOX is methanol inducible, and transcripts driven by this promoter may comprise up to 5% of the total poly(A)+ RNA in induced cells.
P. pastoris has several advantages over Escherichia coli as an expression system, because it does not produce inclusion bodies and it promotes the correct folding of eukaryotic proteins. It also has certain advantages over typical model yeast. First, although its basic N-glycosylation pathway is similar to that of S. cerevisiae and yields glycoproteins with oligomannose-type N-glycans, these structures in P. pastoris have only five to 15 mannose residues (typically Man9GlcNAc2Asn and Man8GlcNAc2Asn) compared with the 50 to 150 mannose residues found in glycoproteins from S. cerevisiae (see Figure 23.2). Hyperglycosylation in S. cerevisiae can interfere with protein folding, requiring use of mnn mutants to limit hyperglycosylation and avoid this problem. Also, P. pastoris does not add outer α1-3-linked mannose residues to its N-glycans. These structures are highly antigenic to humans, making proteins expressed in S. cerevisiae unsuitable for human pharmaceutical use. P. pastoris synthesizes O-glycans with an O-linked mannose core attached to Ser/Thr residues; most of these are short α1-2-linked mannose structures (see Figure 23.3A). Genetic tools have been used to manipulate the machinery for glycoprotein assembly of both S. cerevisiae and P. pastoris. Deletion of ER- and Golgi-specific functionalities in combination with the introduction of heterologous hydrolases and glycosyltransferases has generated “humanized” yeasts and filamentous fungi for the production of therapeutic glycoproteins (Chapter 56). P. pastoris has emerged as a potent orthogonal host to produce exogenous polysaccharides such as those occurring in the plant cell wall.
Kluyveromyces lactis in Industry
Kluyveromyces lactis metabolizes lactose to lactic acid and, along with Aspergillus niger and E. coli, is grown to produce rennet for making cheese and other products. K. lactis is also a rich source of β-galactosidase, which hydrolyzes lactose. K. lactis synthesizes mannans similar to those in S. cerevisiae, but they lack mannose phosphate modifications and some side chains are capped with a residue of N-acetylglucosamine. A K. lactis mutant that lacks these N-acetylglucosamine residues because of a deficiency of the Golgi UDP-GlcNAc nucleotide sugar transporter has been productively exploited in studies of heterologous transporters.
BASIDIOMYCETE DIVERSITY
To convey the enormous diversity of fungi we will consider the basidiomycete phylum, which encompasses fungi that produce spores from a pedestal-like structure called the basidium. Basidiomycetes range from fungi with gills or pores, such as common mushrooms and bracket fungi, to budding yeast that are deadly human pathogens.
Lifestyle and Polysaccharides
The basidiomycete Dictyonema glabratum illustrates a distinct fungal lifestyle. It lives in symbiosis with cyanobacteria Scytonema sp. forming a lichen, which is notable for its many unusual glycans. For example, although the β-glucans of most lichens are linear, in D. glabratum they are branched with β1-3 and β1-6 linkages. The mannans of D. glabratum also have an α1-3-linked backbone, rather than the typical α1-6 linkages found in other lichens, along with branches at the 2 and 4 positions. Finally, the xylans of this organism are linear β1-4-linked polymers of xylose, more typical of those found in “higher” plants and algae than in fungi. D. glabratum also synthesizes several unusual short glycolipids, including glycosyldiacylglycerolipids, which are similar to plant glycolipids and contain monosaccharides, disaccharides, and linear trisaccharides of α1-6-linked galactopyranose. This fungus thus highlights the extensive glycan diversity of the fungal kingdom.
O-Glycans
Another example of glycan diversity is offered by Cryptococcus laurentii, which has the unusual property of producing toxins that kill a pathogenic yeast, C. albicans (see below). The O-glycans of C. laurentii are unusual in that they contain mannose, xylose, and galactose (Figure 23.3C); these glycans are synthesized by a unique set of mannosyl-, xylosyl-, and galactosyltransferases that are not homologous to human enzymes. This diversity would not have been predicted by studies of model yeast alone, emphasizing the importance of examining glycans in a wide range of fungal species.
N-Glycan Diversity
Coprinopsis cinerea is a mushroom species that has gained prominence as a model for studies of diverse topics including mating, sexual development, meiosis, and the evolution of multicellularity. Its N-glycans typically are high mannose type with five to nine mannoses, but may also have a bisecting α1-4GlcNAc at the β-mannose.
PATHOGENIC FUNGI
Pathogenic fungi are a significant cause of plant and animal disease, responsible for devastation of crops, decimation of animal populations (e.g., certain bat, amphibian, and bee species), and serious human diseases that kill an estimated one million people each year. These organisms display diverse glycans, which typically differ from those of the host and have been implicated in multiple pathogenic processes and host–pathogen interactions. For example, during plant infections plant glycosylhydrolases may partially digest wall glucans of invading fungi. Some of the released oligosaccharides, termed oligosaccharins, can then act as signals to promote plant antifungal defenses.
Candida albicans Glycans Are Central in Host Interactions
C. albicans (an ascomycete) is a normal commensal organism that can cause illness ranging from irritations of mucosal surfaces to life-threatening systemic infections. The C. albicans cell wall contains β1-3- and β1-6-linked glucans and chitin, similar to the S. cerevisiae wall, and immunogenic mannans that are termed phosphopeptidomannans. It also produces unusual short β1-2-linked mannose chains (Figure 23.2) that are highly antigenic and are also expressed on phospholipomannan (PLM) antigens. PLM antigens contain phytoceramide derivatives of myo-inositol phosphate. The β1-2 mannosides are linked via an α-mannosylphosphate to the common glycosphingolipid Manα-1,2 inositolphosphoceramide. The abundant GPI-anchored proteins of C. albicans have been implicated in fungal adherence to host tissues.
The O-glycans of C. albicans are short chains of α1-2-linked mannose (Figure 23.3A), which lack the α1-3-linked mannose caps found in S. cerevisiae. As in S. cerevisiae, deficiencies of O-mannose addition generated through genetic deletions are lethal, indicating that O-mannosylation is essential in this yeast. C. albicans mannans are also important in its interactions with host cells, including macrophages and dendritic cells. In particular, these structures are recognized by the mannose receptor and by dectin-2. These are C-type lectins expressed by immune cells that are important in both innate and adaptive immune responses (see Chapter 34). PLM antigens may be shed by C. albicans and, through interactions with Toll-like receptors (TLR-2), they can induce nuclear factor-κB (NF-κB) activation and cytokine responses such as tumor necrosis factor-α (TNF-α) secretion. Galectin-3, a ubiquitous member of the galectin family of lectins that is highly expressed in macrophages, also appears to recognize C. albicans expressing β1-2-linked mannose residues, resulting in opsonization of the yeast.
Aspergillus fumigatus
A. fumigatus is an environmental mold that spreads by airborne particles. It causes serious invasive disease in immunocompromised people that is difficult to treat, leading to high mortality rates. As with other fungal pathogens, the surface glycans of A. fumigatus are critical for interactions with the host. The cell wall of infectious forms of this fungus is covered with specific proteins and melanin, presumably to alter surface properties and mask these structures from recognition by host immune receptors. The hyphal wall has a core of branched β1-3-glucan covalently linked to other glucan components, chitin, and galactomannan, which consists of a mannose backbone with short galactofuranose side chains. Interestingly, galactomannan also occurs anchored to the plasma membrane by a GPI. This polysaccharide is assembled in the Golgi apparatus and is probably transferred to the cell wall by transglycosidases, in the same way as GPI-anchored proteins. A. fumigatus also produces an extracellular matrix composed of monosaccharides, α1-3-glucan, galactomannan, and a galactosaminogalactan composed of variable galactopyranose repeats linked to N-acetylgalactosamine, which are partially de-acetylated. This structure has been implicated in adhesion and fungal virulence. The extracellular matrix also plays an important role in concealing the immunogenic β1-3 glucan layer from the immune system.
Cryptococcus neoformans and Its Capsule
C. neoformans is a ubiquitous environmental basidiomycete yeast that causes severe disease in immunocompromised individuals, leading to roughly half a million deaths per year worldwide. It is unique among pathogenic fungi in having an extensive polysaccharide capsule that is required for virulence (Figure 23.6). The capsule is a dynamic structure that changes in thickness and composition depending on the environment and growth conditions. It is particularly large in the context of mammalian infection, in which it impedes host immune responses. It is composed of two large (millions of Da) polysaccharides named for their monosaccharide components: glucuronoxylomannan (GXM) and glucuronoxylomannogalactan (GXMGal). GXM is an extended α1-3 mannan substituted with β1-2Xyl, β1-4Xyl, and β1-2GlcA (Figure 23.7); a subset of the mannose residues are 6-O-acetylated (not shown). The second polymer, GXMGal, is based on α1-6 galactan, with side chains of galactose, glucuronic acid, mannose, and xylose (Figure 23.7); the backbone is also modified with small amounts of β1-2-linked galactofuranose (not shown). Association of the capsule with the cell surface relies on a cell-wall component, α1-3 glucan. Although α1-3 glucan is not present in the cell walls of S. cerevisiae or C. albicans, it is common in other fungi. N-glycans of C. neoformans are generally high mannose with modest outer chain extensions and may include xylose β1-2-linked to the trimannosyl core. Both N- and O-glycans may also be modified with xylose and xylose phosphate.

FIGURE 23.6.
A quick-freeze deep-etch image of the edge of a Cryptococcus neoformans cell. The polysaccharide capsule (open meshwork at right) is linked to the cell wall (central structure dividing the image from upper left to lower right) via α1-3 glucan. (more...)
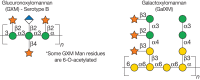
FIGURE 23.7.
Structures of capsular polysaccharides in Cryptococcus neoformans.
Fungal Glycans as Drug Targets
In the context of fungal diseases, the similarity of fungi to their eukaryote hosts becomes a liability, because it is challenging to develop antifungal drugs that are not hampered by toxicity. The unique features of fungal glycans may suggest drug targets to help improve this picture, and decrease the roughly one million deaths each year caused by fungal infections.
The major success story using this approach is the development of echinocandin drugs. These antifungal lipopeptides inhibit β1-3-glucan synthesis in fungi including Candida and Aspergillus, leading to cell wall compromise, and are used clinically to treat invasive fungal infections, although they are not effective against all fungal pathogens. GPI-synthesis inhibitors also offer promise for treatment of fungal pathogens; compounds that selectively inhibit yeast inositol acylation are currently in clinical trials. Continued efforts to target novel aspects of fungal glycobiology may advance the ongoing search for new therapies.
ACKNOWLEDGMENTS
The authors appreciate helpful comments and suggestions from Anne Imberty and Markus Pauly.
FURTHER READING
- Ballou CE, Lipke PN, Raschke WC. 1974. Structure and immunochemistry of the cell wall mannans from Saccharomyces chevalieri, Saccharomyces italicus, Saccharomyces diastaticus, and Saccharomyces carlsbergensis. J Bacteriol 117: 461–467. doi:10.1128/jb.117.2.461-467.1974 [PMC free article: PMC285535] [PubMed: 4590470] [CrossRef]
- Huffaker TC, Robbins PW. 1983. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci 80: 7466–7470. doi:10.1073/pnas.80.24.7466 [PMC free article: PMC389972] [PubMed: 6369318] [CrossRef]
- Dickson RC, Lester RL. 1999. Yeast sphingolipids. Biochim Biophys Acta 1426: 347–357. doi:10.1016/s0304-4165(98)00135-4 [PubMed: 9878820] [CrossRef]
- Poulain D, Jouault T. 2004. Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr Opin Microbiol 7: 342–349. doi:10.1016/j.mib.2004.06.011 [PubMed: 15358252] [CrossRef]
- Daly R, Hearn MT. 2005. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit 18: 119–138. doi:10.1002/jmr.687 [PubMed: 15565717] [CrossRef]
- Klis FM, Ram AF, De Groot PW. 2007. A molecular and genomic view of the fungal cell wall. In Biology of the fungal cell (ed. Howard RJ, Gow NAR, editors. ), 2nd ed, The Mycota VIII, pp. 97–120. Springer-Verlag, Berlin. doi:10.1007/978-3-540-70618-2_4 [CrossRef]
- Deshpande N, Wilkins MR, Packer N, Nevalainen H. 2008. Protein glycosylation pathways in filamentous fungi. Glycobiology 18: 626–637. doi:10.1093/glycob/cwn044 [PubMed: 18504293] [CrossRef]
- De Pourcq K, De Schutter K, Callewaert N. 2010. Engineering of glycosylation in yeast and other fungi: current state and perspectives. Appl Microbiol Biotechnol 87: 1617–1631. doi:10.1007/s00253-010-2721-1 [PubMed: 20585772] [CrossRef]
- Everest-Dass AV, Jin D, Thaysen-Andersen M, Nevalainen H, Kolarich D, Packer NH. 2012. Comparative structural analysis of the glycosylation of salivary and buccal cell proteins: innate protection against infection by Candida albicans. Glycobiology 22: 1465–1479. doi:10.1093/glycob/cws112 [PubMed: 22833316] [CrossRef]
- Loza LC, Doering TL. 2021. Glycans of the pathogenic yeast Cryptococcus neoformans and related opportunities for therapeutic advances. In Comprehensive glycoscience (ed. Barch Jr J), 2nd ed., Vol. I, pp. 479–506. Elsevier, Amsterdam. doi:10.1016/B978-0-12-819475-1.00079-1 [CrossRef]
- Review Fungi.[Essentials of Glycobiology. 2015]Review Fungi.Doering TL, Cummings RD, Aebi M. Essentials of Glycobiology. 2015
- Review Fungi.[Essentials of Glycobiology. 2009]Review Fungi.Cummings RD, Doering TL. Essentials of Glycobiology. 2009
- Protein glycosylation pathways in filamentous fungi.[Glycobiology. 2008]Protein glycosylation pathways in filamentous fungi.Deshpande N, Wilkins MR, Packer N, Nevalainen H. Glycobiology. 2008 Aug; 18(8):626-37. Epub 2008 May 26.
- Review Mannosylation of fungal glycoconjugates in the Golgi apparatus.[Curr Opin Microbiol. 2014]Review Mannosylation of fungal glycoconjugates in the Golgi apparatus.Fabre E, Hurtaux T, Fradin C. Curr Opin Microbiol. 2014 Aug; 20:103-10. Epub 2014 Jun 13.
- Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi.[BMC Evol Biol. 2008]Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi.Veerappan CS, Avramova Z, Moriyama EN. BMC Evol Biol. 2008 Jul 1; 8:190. Epub 2008 Jul 1.
- Fungi - Essentials of GlycobiologyFungi - Essentials of Glycobiology
Your browsing activity is empty.
Activity recording is turned off.
See more...
