The content of this book is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 Unported license. To view the terms and conditions of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology [Internet]. 4th edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. doi: 10.1101/glycobiology.4e.21
Glycoconjugates are integral components of the cell surfaces of bacteria and are often the immediate point of contact with the environment. Surface glycoconjugates contribute to the essential permeability barrier properties of the cell envelope, influence both the susceptibility and resistance of bacteria to antibiotics and other harmful compounds, participate in the formation and dispersion of biofilms, act as receptors for bacteriophages, and play pivotal roles in pathogenic and symbiotic host–microbe interactions. Reflecting these many functions, surface glycoconjugates are remarkably diverse, enabled by the propensity for bacterial genetic recombination and lateral gene transfer and shaped by environmental interactions that impart niche-specific selective pressures. Surface glycoconjugates drive a variety of important interactions with host innate and adaptive immune defenses. Some are recognized as pathogen-associated molecular patterns (PAMPs), for example, via Toll-like receptor (TLR)-mediated pathways. Others are natural targets of adaptive immunity and have been exploited in successful vaccine strategies. Because of their importance in cell viability, surface glycoconjugates are also frequent targets of antimicrobial strategies. This chapter will provide an overview of the structure and biosynthesis of glycoconjugates, together with some examples of their functions.
AN OVERVIEW OF CELL ENVELOPE ARCHITECTURES
Bacteria were historically divided into two major groups based on their response in the Gram-staining procedure (i.e., Gram-positive and Gram-negative organisms), reflecting the organization of the cell wall. The amount and location of peptidoglycan in the cell wall is an important contributor to the outcome of Gram staining. Peptidoglycan is essential for the viability of most bacteria. It consists of polysaccharide strands covalently cross-linked by short peptides, creating a three-dimensional structure that confers shape and rigidity to the cell. In Gram-negative bacteria, such as Escherichia coli, the cell wall consists of two membranes separated by a cellular compartment termed the periplasm in which thin layers of peptidoglycan reside (Figure 21.1). In Gram-positive bacteria, substantially thicker layers of peptidoglycan surround a single membrane and provide an attachment point for other glycan structures. Another class of bacteria, the Negativicutes, are related to the Gram-positive Firmicutes but nevertheless have a two-membrane architecture and stain as Gram-negative. Other bacteria produce variable responses in the Gram-staining reaction, largely because of the presence of other glycans and cell wall lipids (see below).

FIGURE 21.1.
Conceptual organization of the cell envelopes of Gram-negative bacteria, Gram-positive bacteria, and mycobacteria. The schematic comparison of the cell walls (or cell envelopes) of Gram-negative bacteria, Gram-positive bacteria, and mycobacteria illustrates (more...)
The periplasm of Gram-negative bacteria contains proteins associated with cell-surface assembly and nutrient uptake but may also contain free oligosaccharides (fOS) that protect against osmotic stress. The outer membrane is an asymmetric lipid bilayer, with an outer leaflet composed mainly of a unique glycolipid called lipopolysaccharide (LPS) that is essential for the integrity of the permeability barrier imposed by the outer membrane. Many Gram-negative bacteria are covered in a surface-bound polysaccharide layer known as a capsule, and, in some cases, this capsular polysaccharide (CPS) is released from the cell in large amounts as free exopolysaccharide (EPS). Bacteria producing these products are often readily identified by their highly mucoid colonies. Structural variations in the LPS and capsules of different bacterial species are diverse and influence many types of interactions between bacteria and their environmental and host niches.
Gram-positive bacteria lack the outer membrane (Figure 21.1) and depend on a much thicker multilayered peptidoglycan layer for viability. The Gram-positive cell wall is modified with additional specialized cell wall glycan polymers covalently linked to peptidoglycan (e.g., wall teichoic acids [WTAs]), whereas glycolipids, such as lipoteichoic acids (LTAs), are anchored in the cell membrance. Mycobacteria and related Actinobacteria are not considered to be Gram-positive or Gram-negative, and are instead classified as acid-fast bacteria because they possess distinctive cell walls that confer an aberrant response to the classical Gram-staining procedure. Their complex cell walls have a high content of remarkable glycan and glycolipid structures (Figure 21.1). The unique arabinogalactan (AG) component that characterizes these organisms is covalently attached to peptidoglycan and provides an attachment point for characteristic long-chain (C60–C90) α-alkyl-β-hydroxymycolic acids. The resulting mycolyl-AG-peptidoglycan complex underpins a permeability barrier conferring resistance to antibiotics and other harmful molecules. Mycolic acids help form an envelope layer referred to as an “outer membrane,” although its structure shares no similarity with its namesake in Gram-negative bacteria.
PEPTIDOGLYCAN, A DYNAMIC STRESS-BEARING LAYER
Structure, Arrangement, and Function
Peptidoglycan (also known as murein) forms a rigid sacculus enveloping the cytoplasmic membrane, conferring cell shape, and contributing to the ability of bacteria to resist the effects of internal osmotic (turgor) pressure. Although most bacteria possess peptidoglycan, it is absent in some obligate intracellular pathogens, such as Mycoplasma sp., and can be conditionally expendable in certain bacteria that produce L-forms. Given its distribution, peptidoglycan is recognized by host defenses as a marker of infection. Released peptidoglycan is a PAMP recognized by TLR2 and pattern recognition proteins, such as nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), to activate an inflammatory response as part of innate immunity.
Peptidoglycan makes up ∼10% of the dry weight of the cell wall in Gram-negative bacteria, in which it exists in a structure one to three layers thick. In contrast, Gram-positive bacteria have a thicker cell wall containing 10 to 20 layers of peptidoglycan, representing as much as 20%–25% of the dry weight of the cell. The overall chemical structure of peptidoglycan is similar across bacterial genera (Figure 21.2) and consists of parallel strands of polysaccharides composed of a β1-4-linked disaccharide of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc). The average chain length of the glycan strands in E. coli is 25–35 disaccharide units but may increase several-fold in Gram-positive bacteria. Adjacent strands are connected to one another via cross-links between short peptide strands attached to the MurNAc residues (Figure 21.2). Individual glycan strands are thought to be arranged parallel to the membrane and the helical conformation of the glycan strands enables cross-linking in three dimensions, within and between layers, to create the functional higher-order structure. Although the amino acid composition varies in different bacteria, the peptides are typically composed of L-Ala, unusual D-amino acids (e.g., D-Glu and D-Ala) and a dibasic amino acid, such as meso-diaminopimelic acid (m-A2pm or m-DAP) or L-Lys, to facilitate the cross-linking. Cross-links may be direct peptide bonds or (in some Gram-positive bacteria) include several amino acids, such as Gly5. This, and the frequency of cross-linking, will have spatial implications for the final meshwork. Variations in peptidoglycan structure may occur within the same organism at different stages of the cell cycle, during developmental processes, or at different sites in the same cell.

FIGURE 21.2.
Structure, biosynthesis, and inhibition of peptidoglycan assembly. Peptidoglycan creates a meshwork composed of glycan chains with a disaccharide unit containing β1-4-linked N-acetylglucosamine and N-acetylmuramic acid (MurNAc, purple; GlcNAc, (more...)
A characteristic property of many Gram-positive cell walls is the attachment of secondary cell wall polymers to MurNAc residues in the glycan backbone. In addition, the dibasic amino acid provides a potential site for protein attachment. Sortase-family enzymes link a wide range of proteins (including pili, surface-layer [S-layer] subunits, and metabolite-binding proteins) to the cell wall. In Gram-negative bacteria, outer membrane lipoproteins are attached to this site in peptidoglycan.
Despite its role in cell integrity, peptidoglycan is a dynamic structure, allowing insertion of new material during rapid cell growth and division, as well as accommodating the assembly of macromolecular structures (e.g., flagella and protein secretion systems), which pass through the cell wall. As a result, peptidoglycan undergoes ∼50% turnover every generation, catalyzed by a collection of peptidoglycan hydrolases (glycosidases, peptidases, and amidases). An imbalance in the metabolism of peptidoglycan can lead to a rapid loss of cell wall integrity, followed by osmotic swelling and cell lysis. Such effects underpin the actions of cell wall–active antibiotics (described below) and the effect of lysozyme, an enzyme secreted by leukocytes as part of innate immunity.
Assembling the Cell Wall
Peptidoglycan synthesis involves a complex series of well-conserved enzymatic reactions localized to three cellular compartments (Figure 21.2). In the initial stage, activated nucleotide precursors are synthesized in the cytoplasm. UDP-MurNAc is generated through the condensation of phosphoenolpyruvate (PEP) with UDP-GlcNAc and subsequent reduction. In some mycobacteria UDP-MurNAc is hydroxylated into UDP-MurNGc for unknown reasons, generating the only other known N-glycolyl group in nature (besides N-glycolylneuraminic acid, which is exclusive to deuterostomes) (Chapter 15). Sequential addition of the amino acids by specific ATP-dependent amino acid ligases leads to the formation of the UDP-MurNAc-pentapeptide. The disaccharide repeat unit is built on a C55 polyprenol carrier (undecaprenol or bactoprenol phosphate) in the cytoplasmic membrane in a process resembling the early stage of eukaryotic N-linked glycan formation (Chapter 9). The resulting lipid-linked disaccharide subunit (lipid II) is flipped across the membrane mainly by MurJ, although other flippases may also participate in some circumstances. Lipid II is polymerized at the periplasmic face of the membrane and new glycans are inserted into the growing cell wall. This requires glycosyltransferases (GTs) to extend the glycan backbones and transpeptidases (TPs) to catalyze cross-linking. Each organism possesses multiple GT and TP activities, existing as monofunctional enzymes or as bifunctional GT-TPs, and structures of key enzymes have provided insight into their mechanisms. GTs mediate stepwise assembly of the glycan using a molecule of lipid II as the acceptor and releasing one undecaprenyl diphosphate in each cycle.
The TP and GT reactions are context-dependent; active complexes containing different enzyme combinations are coupled with peptidoglycan remodeling enzymes that are guided by cytoskeletal elements (actin and tubulin homologs) to achieve wall elongation or cell division. This process is an essential driver of cell shape determination. Division is particularly complex, with a multiprotein “divisome” that includes regulatory proteins dedicated to fidelity in the timing and placement of the division site. Among the remodeling enzymes are lytic transglycosylases, which cleave glycan chains to create a characteristic terminal 1,6-anhydroMurNAc residue. This reaction may determine the size of the glycan chains (effectively marking the end of biosynthesis) but is also required for the insertion of trans–cell wall structures, signaling in the regulation of some β-lactamases, and the release of PAMPs.
Antibacterials Targeting Peptidoglycan Assembly
Peptidoglycan synthesis has offered a classic target for antibiotics and continues to provide new avenues for drug discovery. Penicillins and other β-lactam antibiotics mimic the D-Ala-D-Ala motif and create a slow-turnover acyl-enzyme complex with TPs (hence, proteins with TP modules are also referred to as penicillin-binding proteins, or PBPs). The action of these drugs requires active bacterial replication. Resistance to these compounds involves their exclusion from the cell, replacement of the target enzyme, or production of β-lactamases. Vancomycin and other glycopeptides also affect the cross-linking step by binding to the terminal D-Ala-D-Ala to prevent its turnover. Members of the moenomycin family of compounds are phosphoglycolipids that are thought to inhibit transglycosylase GTs by mimicking the growing glycan chain and binding the donor substrate site in the enzyme. The points of action of these and other antibacterial compounds are shown in Figure 21.2.
GRAM-POSITIVE BACTERIA PRODUCE ADDITIONAL CELL WALL GLYCOPOLYMERS
Gram-positive cell walls contain large amounts of additional polymers that provide further functionality and structure to the cell wall. The best known examples are WTAs, which often consist of polyribitol or polyglycerol chains (40 to 60 repeats) variably substituted with monosaccharides and/or D-Ala. These chains are linked by a phosphodiester bond to the C6 position of MurNAc through a conserved linkage region (Figure 21.3). WTAs extend through and beyond the peptidoglycan layers and are exposed at the cell surface. They are expendable for viability in the laboratory, yet are required for pathogenicity and potentially for growth in other environments. A wide range of functions are (directly or indirectly) attributed to WTAs, including regulation of peptidoglycan remodeling, cell wall growth and morphology, binding divalent cations, adhesion, biofilm formation, and resistance to innate immunity (lysozyme and antimicrobial peptides), as well as resistance to antibiotics and other environmental stresses. WTAs also provide a means of tethering some proteins, including S-layer proteins, to the cell surface. Under conditions of phosphate starvation, some bacteria replace the phosphate-rich WTAs by teichuronic acids, where carboxyl groups create their anionic character (Figure 21.3).

FIGURE 21.3.
Structures of additional cell wall polymers in classical Gram-positive bacteria and mycobacteria. A range of additional (secondary) wall polymers extend the structure and functionality of peptidoglycan. Wall teichoic acids (WTAs) typically contain a polyol-phosphate (more...)
WTAs are synthesized from nucleotide-activated precursors on undecaprenyl carriers and, once complete, are exported across the cytoplasmic membrane by an ATP-binding cassette (ABC) transporter (like LPS O-PS, see below) and attached to MurNAc acceptors. The timing (during peptidoglycan assembly) and location of WTA insertion is not fully resolved.
LTAs are also commonly found in Gram-positive cell envelopes. Some LTAs may possess similar carbohydrate structures to WTAs (Figure 21.3), whereas others are substantially more complex. They are attached to membrane anchor glycolipids that vary in sugar composition but typically contain terminal diacylglycerol (DAG). The assembly pathways of WTAs and LTAs are quite different. For example, the prevalent polyglycerol-based LTAs require phosphatidylglycerol as the glycerol donor, rather than a nucleotide-activated precursor seen in WTA synthesis, and LTAs are assembled on the DAG-containing glycolipid without the participation of undecaprenyl phosphate. LTAs may coexist with WTAs, and some of their proposed functions overlap, although LTAs may be of particular importance in those bacteria that do not contain teichoic acids. Mutations eliminating both LTA and WTA synthesis show synthetic lethality. LTAs have also been implicated in inflammatory responses and are proposed as ligands for a range of host receptors, although information concerning their status as PAMPs recognized by TLR2 is contradictory.
MYCOBACTERIA POSSESS UNUSUALLY COMPLEX CELL ENVELOPE GLYCOCONJUGATES
Pathogenic mycobacteria, including the organisms that cause tuberculosis and leprosy, are intracellular parasites that replicate within modified phagosomes of macrophages. Mycobacteria are also able to enter a dormant (metabolically inactive) state, aiding their persistence. These properties are critically dependent on a remarkable cell envelope architecture based on a wide array of unusual cell wall constituents (Figure 21.1). AG is crucial to the integrity of the cell wall and the organism's resistance to many antibiotics. AG is anchored to approximately one in every 10 disaccharides in a peptidoglycan strand, in a similar manner as WTAs (Figure 21.3). The main carbohydrate poly-Galf backbone of AG is synthesized on a decaprenyl-P carrier in a process resembling some LPS O-antigens (see below), before its export to the periplasm, which is proposed to involve a transporter. Elaboration of the arabinofuran is performed in the periplasm, using decaprenyl-P-Araf as the immediate donor. After ligation to peptidoglycan, the reducing terminal Araf residues provide attachment sites for the characteristic mycolic acids. The lipids form a major part of the mycobacterial outer membrane (Figure 21.1). This unusual outer membrane offers a formidable barrier against antibiotics and harmful compounds. The essential role of the mycolyl-AG-peptidoglycan complex in mycobacterial physiology makes it an attractive target for drug discovery. Indeed, the frontline drug ethambutol is a structural analog inhibitor of arabinosyl GTs.
The cell envelope also contains a wide range of bioactive glycolipids. Abundant examples are based on a core structure of phosphatidylinositol mannosides (PIMs), with varying acylation and glycan lengths. PIMs and PI can account for >50% of the total envelope phospholipid content and are important for cell wall integrity and division. After synthesis and export, these lipids are extended into lipomannans (LMs) by additions from decaprenyl-P-Man donors. As is the case for AG, periplasmic extension of LMs is catalyzed by specific type C GTs that further elaborate into lipoarabinomannans (LAMs) using decaprenyl-P-Araf, creating a covalently linked arabinan resembling the structure in AG, in a LAM molecule containing more than 100 sugars (Figure 21.3). The terminal Araf residues can be further capped by Man residues in ManLAM. These compounds are all potentially recognized by an adaptive immune response, the mannose receptor on macrophages (facilitating phagocytosis), the lectin pathway of complement activation, and by DC-SIGN (dendritic cell–specific intercellular adhesion molecule-3-grabbing nonintegrin). ManLAM and (particularly) LM are potent ligands for TLR2 but differ in their immunomodulatory effects. Indeed, their exact role(s) in the complex pathogenesis process remain unresolved.
LIPOPOLYSACCHARIDE (ENDOTOXIN)—A KEY COMPONENT IN MOST GRAM-NEGATIVE BACTERIA
The outer membrane of Gram-negative bacteria (Figure 21.1) is an asymmetric bilayer, with the outer leaflet that is mostly composed of the unique glycolipid known as LPS (see Figure 21.5). LPS is absent in only a few Gram-negative bacteria, such as the endosymbionts Borrelia and Wolbachia and in Sphingomonas, in which LPS is functionally replaced by sphingolipids. In Salmonella, there are ∼106 molecules of LPS per cell (vs. 107 total phospholipids), covering ∼75% of the cell surface. LPS is usually essential for outer membrane integrity in producing organisms. However, a few species (e.g., Neisseria meningitidis and Acinetobacter baumannii) survive in the face of mutations that prevent LPS synthesis, although this imparts a cost on fitness and virulence. Much of our knowledge of the structure, synthesis, and functions of LPS arises from work in E. coli and Salmonella, but the general principles are widely applicable. LPS consists of three structural domains: lipid A, core oligosaccharide, and the O-antigen polysaccharide (O-PS) (Figure 21.4). Some mucosal pathogens, such as Neisseria and Campylobacter, produce a truncated (nonrepeating) glycan that may be subject to phase variation. These LPS forms are often referred to as lipooligosaccharide (LOS) (Figure 21.4).

FIGURE 21.4.
Structural organization of lipopolysaccharides (LPSs). LPSs are characteristic components of Gram-negative outer membranes, and the cartoon in A illustrates different formats of LPS structures. LPS composed of only the lipid moiety (lipid A) is rare in (more...)
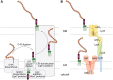
FIGURE 21.5.
Assembly and export of lipopolysaccharides (LPSs). The complexity in LPS structures is reflected in the assembly pathways (A). Lipid A is assembled at the cytosol cytoplasmic membrane (CM) interface by proteins designated Lpx* (where the asterisk marks (more...)
Structure and Function
Lipid A is a glycolipid containing a variable number of fatty acyl chains covalently attached to a disaccharide backbone that anchors LPS in the outer membrane. The most common lipid-A structure consists of a β1-6-linked diglucosamine backbone. In E. coli, the reducing terminal sugar contains phosphate at C-1 and ester- and amide-linked β-hydroxymyristic acid residues at C-3 and C-2, respectively (Figure 21.4). The second glucosamine residue also contains phosphate at C-4′ and two β-hydroxymyristic acids, in ester linkage at C-3′ and amide linkage at C-2′, which carry additional lauroyl groups on their β-hydroxyls. LPS was first discovered in the late 1800s as a heat-stable toxin associated with bacteria, and chemical synthesis of E. coli lipid A in 1985 confirmed that this moiety is responsible for the biological (endotoxic) properties of LPS in mammals, resulting in fever, septic shock, and other deleterious physiological effects (Chapter 42). When released from the cell surface, lipid A is a PAMP recognized by the TLR4/myeloid differentiation factor 2 (MD2) receptor, which dimerizes to trigger secretion of proinflammatory mediators. This can result in beneficial stimulation of the adaptive immune response and has been used as a vaccine adjuvant (i.e., monophosphoryl lipid A). However, LPS-mediated inflammation can also cause morbidity and mortality and some bacteria (e.g., commensals of the gut microbiome) have adapted LPS chemistries that minimize inflammation. In some Gram-negative species, these different outcomes are dictated by variable phosphorylation and acylation of the lipid-A structure, which are subject to environmental regulation and result in altered signaling downstream from TLR4/MD2.
Free lipid A occurs in a few rare exceptions (e.g., Francisella novicida) because, in most bacteria, lipid A is modified by sugars forming the core oligosaccharide. This branched oligosaccharide is conceptually separated into a more variable outer core that provides an attachment point for the O-antigen, whereas the inner core is more conserved. All lipid-A molecules contain one to four units of the unusual eight-carbon sugar, Kdo (3-deoxy-D-manno-oct-2-ulosonic acid [or a derivative thereof]), located at the linkage region between lipid A and core, and many inner cores contain L (or D)-glycero-manno-heptose residues. Negatively-charged components in the inner core, such as phosphates and uronic acids, provide further binding sites for the divalent cations that stabilize the outer membrane. The outer core is more variable; for example, five different structures are found in different isolates of E. coli.
The outermost portion of LPS is the hypervariable O-PS, which forms the O-antigen used in serotyping clinical isolates (e.g., E. coli O157). There are more than 180 structurally and serologically distinct O-antigens in E. coli alone and these arise from recombination and horizontal gene transfer events and selection pressures, including host immune response and O-PS specific bacteriophages. This diversity reflects a remarkable range of sugar residues, including free and amidated uronic acids, amino sugars, methylated and deoxygenated derivatives, acetylated sugars, and nonsugar substituents (e.g., amino acids and phosphate). O-PSs are repetitive polysaccharides with one to eight sugars (and noncarbohydrate substituents) per repeat unit. Each chain can contain one to hundreds of repeats, although there is a preferred (modal) cluster of chain lengths in LPS isolated from an individual strain. This is evident in the characteristic cluster of bands in the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) profile (Figure 21.4), where each higher band reflects an LPS molecule containing an O-PS with one additional repeat unit. In the mammalian gut, LPSs are targets of multivalent IgA that aggregates growing bacteria, leading to entrapped growth and prevention of (pathogen) spread. In turn, the high variability and phase-variation of LPS structures or phage-mediated LPS remodeling may be outcomes of selection in the face of adaptive immunity. In some mammalian pathogens, O-PS chain length is critical for resistance to complement-mediated killing, but it can also influence interactions of bacteria with macrophages. Other mammalian pathogens that synthesize (nonpolymeric) LOS incorporate structural variations that enable resistance to one or more arms of the complement system. The exposure of O-PSs makes them prime targets as receptors for bacteriophages, and this may be a driving force in the evolution of their structural diversity.
The Complex Path for Assembly and Export of LPS
Substantial effort has been invested in studying LPS biosynthesis and in developing approaches to target the enzymes involved in LPS formation. Lipid A biosynthesis involves a highly conserved nine-step pathway (the Raetz pathway), named after Christian Raetz, whose research group was largely responsible for its discovery and characterization. In the E. coli prototype, it begins with the precursor UDP-GlcNAc and culminates with the product lipid A-Kdo2. The early steps in this pathway occur in the cytosol, whereas the acylation steps are membrane-associated. The two β-hydroxyl-linked fatty acids are added only after the Kdo units. As such, mutations (or inhibitory compounds) affecting the precursor CMP-Kdo block lipid A synthesis at the stage of the tetraacyl (lipid IVA) intermediate. The formation of Kdo2–lipid A is normally essential for survival of E. coli, but this does not hold true in a few bacteria (including N. meningitidis and A. baumannii). Once complete, lipid A-Kdo2 molecules may enter the LPS export pathway or provide an acceptor for addition of the core oligosaccharide by the sequential actions of nucleotide sugar-dependent GTs and kinases before export. Lipid A (or lipid A-core) are substrates for MsbA, an ABC transporter that flips these molecules to the periplasm.
At the periplasmic face, lipid A-core molecules may be modified by the addition of O-PS or translocated directly to the cell surface (Figure 21.5), contributing to heterogeneity in the final LPS species that is reflected in the SDS-PAGE profile (Figure 21.4). The assembly of O-PS occurs independently of the lipid A-core, with most bacterial species using one of two main synthetic strategies. Both strategies involve the formation of undecaprenyl diphosphate-linked intermediates at the cytoplasmic face of the inner membrane, using nucleotide sugars as activated donors. In one pathway, individual lipid-linked repeating units are synthesized and flipped to the periplasmic face of the membrane by a transporter (Wzx), related to MurJ from peptidoglycan assembly (see above). In the periplasm, the polymerase (Wzy) transfers a nascent O-PS chain from its undecaprenol carrier to the nonreducing terminus of the newly exported lipid-linked repeating unit, extending the chain one repeat unit at a time. An additional protein, Wzz (a member of the polysaccharide copolymerase [PCP] family), regulates the polymerization reaction to yield O-PS chains within a particular (modal) size range that is appropriate for their function(s). This generates the distribution seen in SDS-PAGE profiles (Figure 21.4). In the alternative pathway, the full-length O-PS is completed in the cytoplasm by sequential sugar transfer to the nonreducing terminus of the lipid-linked glycan. The final polymerized product is then exported by a dedicated ABC transporter, which shares some structural (and presumably functional) similarity with the WTA transporter. Depending on the bacterial species, the length of the O-PS is determined either by competition between the export and chain-elongation enzymes, or by a chain-capping enzyme. Regardless of which pathway is used, completed undecaprenol diphosphate-linked O-PSs are available in the periplasm, where O-PS ligase (WaaL) completes the glycosylation of the lipid A-core acceptor (Figure 21.5). This enzyme is an integral membrane GT whose catalytic site resides in the periplasm.
Final translocation of LPS molecules to the cell surface is mediated by a conserved LPS translocation pathway comprising seven principle protein components (Figure 21.5). This ATP-driven molecular machine exports an estimated 70,000 molecules/min in growing E. coli. An ABC protein complex (LptBFG) extracts the LPS from the inner membrane and transfers it into the periplasmic part of the pathway. In E. coli, a scaffold comprised of LptA is flanked by domains from LptC and LptD to bridge the two membranes. These domains share the same fold with LptA and are thought to create a unidirectional groove that sequesters the acyl chains in the trafficking LPS molecules. At the outer membrane, the LPS molecules are proposed to enter the lumen of LptD and escape into the membrane via a lateral opening. Ongoing ATP hydrolysis drives the continuing unidirectional export.
PROTEIN GLYCOSYLATION—AN EXPANDING FACET OF BACTERIAL GLYCOBIOLOGY
Protein glycosylation was once thought to be a property confined to eukaryotes. However, it is now apparent that N- and O-linked protein glycosylation systems are also present in Archaea and many genera of bacteria.
Extensive advances have been made in understanding the biosynthesis of bacterial glycoproteins and four general pathways have been identified. There are the traditional pathways for N- and O-linked glycosylation resembling their eukaryotic counterparts and nontraditional pathways that are unique to bacteria. Traditional N-glycosylation systems have only been described in Gram-negative bacteria, with Campylobacter jejuni as the model. Nucleotide-activated sugars in the cytoplasm are assembled onto undecaprenylphosphate in steps resembling the early stages of Wzx/Wzy-dependent O-PS biosynthesis. The completed oligosaccharide is then flipped en bloc across the inner membrane into the periplasm, the topological equivalents of the eukaryotic endoplasmic reticulum membrane and lumen, respectively. Subsequently, the bacterial ortholog of STT3 from the oligosaccharyltransferase (OTase) complex transfers the oligosaccharide posttranslationally onto asparagine residues of at least 80 different proteins by recognizing the bacterial sequon: D/E-X1-N-X2-S/T (where X1 and X2 cannot be proline). The sole OTase protein, PglB, also hydrolyzes the oligosaccharide from the lipid and releases this glycan as fOS into the periplasm. In C. jejuni, fOS concentrations can comprise 2.5% of the dry cell weight and are influenced by osmotic stress. In C. jejuni, a cluster of 10 pgl genes encodes enzymes responsible for the synthesis and transfer of the heptasaccharide: GalNAcα1-4GalNAcα1-4[Glcβ1-3]GalNAcα1-4 GalNAcα1-4GalNAcα1-3-diNAcBac-β1 (diNAcBac is 2,4-diacetamido-2,4,6-trideoxyglucopyranose). The N-glycans of Helicobacter pullorum, Helicobacter winghamensis, Wolinella succinogenes, and Desulfovibrio gigas have also been examined. An active full-length PglB OTase from Campylobacter lari was cocrystallized with an acceptor peptide, providing the first structure of any OTase. The crystal structure of the C. jejuni ABC transporter (i.e., PglK flippase) was also solved and showed that the heptasaccharide and pyrophosphate linker are shielded from the inner membrane lipid bilayer, whereas the undecaprenyl anchor interacts with the lipid bilayer and is responsible for triggering translocation.
γ-Proteobacteria, such as Haemophilus influenzae, Yersinia enterocolitica, and Actinobacillus pleuropneumoniae, possess a nontraditional cytoplasmic pathway for N-glycosylation that does not involve assembly of oligosaccharides on a lipid-linked intermediate, but instead transfers single glycan moieties to proteins from nucleotide-activated precursors. The soluble GTs (NGTs) required for N-glycosylation in these organisms are also completely unrelated to the conventional STT3-orthologous OTases, yet prefer the eukaryotic sequon, N-X-S/T. H. influenzae N-glycosylates its autotransporter adhesion HMW1 with single glucose and galactose residues. In contrast, Shewanella oneidensis, Pseudomonas aeruginosa, and N. meningitidis add rhamnose to Arg32 of their polyproline specific elongation factor to rescue stalled ribosomes.
In addition to N-glycosylation, bacteria possess both canonical and noncanonical pathways for O-glycosylation. For the O-glycosylation system that mirrors the process in eukaryotes, both Gram-positive and Gram-negative bacteria use nucleotide-activated sugars and soluble GTs in the cytoplasm to modify serine and threonine residues on specific proteins, particularly surface structures, such as flagella, pili, and adhesins. Pseudaminic acids, legionaminic acids, and related nonulosonic sugars are common monosaccharides attached by C. jejuni, Helicobacter pylori, and Aeromonas caviae to their flagella and these modifications are essential for filament assembly and bacterial motility.
The noncanonical pathway for O-glycosylation in Gram-negative bacteria involves glycosyltransferase assembly of nucleotide-activated sugars onto undecaprenylphosphate, flipping of the completed oligosaccharide en bloc across the inner membrane into the periplasm, and addition of glycans onto S/T residues by a general OTase. This alternate pathway has been thoroughly characterized in Neisseria species, in which multiple proteins (including pilin) are glycosylated with an oligosaccharide containing diNAcBac (or variants such as glyceramidoacetamidotrideoxyhexose or diacetamidodideoxyglucopyranose) at the reducing end, the same carbohydrate attached to Asn residues by the C. jejuni N-glycosylation pathway. Gram-positive bacteria lack surface LPS, but many instead possess a regular array of glycoproteins known as S-layer proteins. The oligosaccharides on these S-layer proteins are also first assembled onto an undecaprenylphosphate carrier and subsequently added onto S/T residues of these proteins, but can also be coupled to hydroxyl groups of tyrosine residues.
OSMOREGULATED PERIPLASMIC GLUCANS
Bacteria encounter extreme differences in osmolarity in the environment (and must withstand up to 6 atmospheres of turgor pressure!) and have thus evolved both physical and chemical mechanisms to resist disruption. Peptidoglycan, as discussed earlier, provides a structural barrier to osmotic swelling. In many α-, β-, and γ-proteobacteria, a chemical mechanism also exists to protect the inner membrane. In these Gram-negative organisms, osmoregulated periplasmic glucans (OPGs; previously known as membrane-derived oligosaccharides [MDOs]) contribute to an osmotic buffer. These compounds constitute up to 5% of the dry weight of E. coli, and their synthesis is induced by low osmotic conditions. These characteristics are similar to the production of fOS derived from the N-glycosylation pathways of Campylobacter species, which lack normal OPGs.
OPGs were first discovered during analysis of Agrobacterium tumefaciens culture supernatants and rediscovered in studies of phospholipid turnover in E. coli, which showed that the polar head groups of phosphatidylglycerol were transferred to low-molecular-weight, water-soluble oligosaccharides (and thus named MDOs). Other organisms, including Pseudomonas, Rhizobia, Brucella, and Salmonella, also make these compounds. However, because not all oligosaccharides contain membrane phospholipid head groups, they were renamed OPGs. Although the precise structure of OPGs varies, they all consist of D-glucose units with a β-linked backbone. The OPGs comprise four families based on their structure. Family I members, including E. coli, join 5–12 linear glucose units in β1-2 linkage with α1-6 glucose branches. Family II members, including Agrobacterium, synthesize cyclic glucans with 17–25 glucose units joined by β1-2 linkages. Family III/IV members, including Bradyrhizobium, also synthesize cyclic glucans with 10–28 glucose units joined by both α- and β-linkages. In addition, the oligosaccharides can contain phosphoethanolamine, phosphoglycerol, and phosphocholine, as well as acetyl, succinyl, methylmalonyl, and phosphoryl groups, which could add charge to the neutral species.
In E. coli, one inner membrane glucosyltransferase (OpgH), requiring UDP-Glc as a donor and acyl-carrier protein as a cofactor, is involved in the biosynthesis and transport of OPGs across the inner membrane. A second periplasmic glucosyltransferase (OpgG) is involved in glucose branch addition and size regulation of the OPG molecules. Interestingly, OpgH was recently shown to play another role as a nutrient-dependent regulator of E. coli cell size. It is, therefore, not surprising that OPGs influence several biological processes in addition to their involvement in osmoregulation.
EXTRACELLULAR POLYSACCHARIDES
Structure and Function of Capsules and Exopolysaccharides
Bacteria produce long-chain extracellular polysaccharides with extensive structural diversity (Figure 21.6). Extreme examples of this diversity are E. coli, with >80 capsule types, and Streptococcus pneumoniae, with >90 capsule types. There are two types of extracellular polysaccharides. CPSs maintain an association with the cell surface, encapsulating the bacterium in a hydrophilic coating; in contrast, secreted EPSs have limited association with the cell surface postexport.

FIGURE 21.6.
Structures of exopolysaccharides and capsular polysaccharides (CPSs and EPSs). Like O-antigenic polysaccharides, the structures of CPS and EPS are highly diverse, and the main structural features are covered by the examples shown. The structures are divided (more...)
The basis for the retention of CPSs at the cell surface is not always known. In Streptococcus, this is sometimes achieved by covalent linkage to peptidoglycan, similar to other wall-associated polymers, such as teichoic acids (see above). However, streptococcal CPS can also be attached to an unknown membrane lipid. Some E. coli CPSs possess a novel terminal glycolipid composed of an oligosaccharide of several β-linked Kdo residues attached to phosphatidylglycerol, creating a glycolipid with three domains that is conceptually similar to LPS. This terminal glycolipid is conserved in some other Gram-negative mucosal pathogens. In other cases, noncovalent charge-based interactions with other components of the cell surface may be more important.
Extracellular polysaccharides have functions as varied as the environmental niches occupied by the producing bacteria. Given their highly hydrated nature, these polymers are frequently linked to protection against desiccation. Some, such as bacterial cellulose and PNAG (Figure 21.6), are pivotal in biofilm formation. PNAG shows a particularly broad distribution, spanning Gram-positive and -negative species. In many cases, they mask underlying receptors for bacteriophages, although CPSs themselves provide additional receptors and this may help drive the structural diversity. For a bacteriophage to infect encapsulated bacteria, it must penetrate the polysaccharide layer; for this reason, bacteriophages provide a wide range of glycosidases with unique specificities. CPSs are prevalent in pathogens, in which they prevent phagocytosis and (in some cases) complement-mediated killing. Antibodies directed to CPSs overcome these limitations, with the effective pneumococcal vaccines (e.g., Prevnar and Pneumovax) providing excellent examples of the potential of CPSs as vaccine candidates. However, some CPSs in important pathogens are poorly immunogenic because of their mimicry of host cell components. Examples include production of hylauronan by group A streptococci (Chapter 16), α2-3sialyllactosamines by group B streptococci (Chapter 37), α2-8-linked polysialic acid, which mimics embryonic NCAMs, made by E. coli K1 and N. meningitidis (meningococcus) serogroup B, among others, and production of mimics of the glycosaminoglycans (Chapter 17) heparosan and chondroitin by isolates of E. coli and Pasteurella multocida (Figure 21.6). These are remarkable examples of convergent evolution by bacteria, generating molecular mimics of “self-associated molecular patterns” (SAMPs) of eukaryotes.
EPSs may also aid in health promotion. CPS production is vital for the ability of Bacteroides species to colonize the gut, and are the most abundant members of the gut microbiota. In Bacteroides fragilis, a phase-variation mechanism controls the expression of multiple loci (generating multiple structures), but only one is needed for colonization. However, certain CPS structures (e.g., polysaccharide A [PSA], not to be confused with polysialic acids; Figure 21.6), show zwitterionic properties and exert a powerful role in immunomodulation and maturation of the immune system. The capacity of other members of the microbiota and “probiotic” bacteria (e.g., Gram-positive Bifidobacteria) to produce EPSs (Figure 21.6) with wide-ranging physicochemical properties may have additional positive impacts on human health.
EPSs are prominent in a variety of plant pathogens and symbionts. For example, the EPS produced by the Gram-negative phytopathogen Xanthomonas campestris is critical for its detrimental effects on feed crop brassicas. The polymer (known as xanthan gum; Figure 21.6) blocks the flow of water, leading to wilting and further damage. The unusual physical properties of xanthan gum have led to a variety of commercial applications, including as food additives (thickeners). Biofilms are particularly dependent on EPSs. Biofilms are dynamic and structurally complex bacterial communities enclosed within a matrix composed of polysaccharides, proteins, and nucleic acids. They can be found on tissue surfaces and teeth (where they form plaque), as well as on nonbiological surfaces, such as pipelines, ship hulls, and medical implants (including catheters and prostheses). Within the biofilm, a combination of the protected environment and altered cell physiology can help protect bacteria from some antimicrobial compounds. Some bacteria may produce multiple EPSs that contribute to biofilms, and their precise structures may be critical for creation of a proper biofilm architecture, as in Pseudomonas. Certain glycan structures appear well adapted to roles in biofilm communities and are, therefore, more widely distributed. Many biofilm-forming bacteria produce PNAG, also known as polysaccharide intercellular adhesin, with this being targeted by small-molecule inhibitor and vaccine strategies to eradicate biofilm bacteria (Figure 21.6).
Assembly and Export of Capsules and Exopolysaccharides
Assembly systems for CPSs and EPSs are broadly divided into three mechanisms that are distinguished (in part) by the nature of the acceptor on which they are built (Figure 21.6). A large variety of CPSs and EPSs use undecaprenol diphosphate-linked intermediates and follow a synthetic pathway indistinguishable from that used by Wzx/Wzy-dependent O-antigens, described above. In fact, some bacteria produce O-antigens and CPSs/EPSs with the same structures. The CPS shares the same early steps in the pathway but instead of ligating the product to lipid A-core, a CPS-dedicated export machinery spanning the periplasm and outer membrane is used. The features that distinguish CPSs and EPSs in this type of assembly process are unknown. In Gram-positive bacteria with Wzx/Wzy-dependent processes, the nascent polysaccharide is then either released as an EPS or transferred from undecaprenol and attached to peptidoglycam, often via a phosphodiester linkage to MurNAc residues. In Streptococci, this may be catalyzed by an enzyme related to those that link WTAs to the same site in peptidoglycan. Several Gram-negative bacteria assemble CPSs using a conserved phosphatidylglycerol-(Kdo)n acceptor. Although prototypes are found in E. coli and N. meningitidis, the system is conserved across a range of mucosal pathogens. Here, polymerization of the lipid-linked glycan is completed in the cytoplasm, after which it is exported via an ABC transporter. The transporter is linked to additional proteins that span the periplasm and outer membrane in a configuration thought to resemble certain drug efflux pumps in bacteria. The final assembly pathway seems to operate independently of a lipid carrier and requires a processive polymerase called a synthase. Examples are found in Gram-positive and Gram-negative bacteria, but seem to be confined to EPSs. The prototypical hyaluronan and cellulose synthases from Streptococci and Gram-negative bacteria show different architectures, yet both are sufficient for both synthesis and export across the cytoplasmic membrane. The molecular details underlying these processes have been resolved by analysis of the crystal structure of the cellulose synthase. In Gram-negative bacteria, a periplasmic scaffold links the synthase to an outer membrane protein channel.
ADDITIONAL FACETS OF BACTERIAL GLYCOBIOLOGY
Although the focus of this chapter is on the glycobiology of bacterial cell envelopes, there are several additional (and equally important) facets of bacterial glycobiology. Three of these are briefly mentioned here. First, in addition to synthesizing surface structures, bacteria also produce intracellular glycans and cytoplasmic glycosylated proteins and glycolipids. For example, glycogen and trehalose act as storage compounds within some bacteria. Mycobacteria also contain a growing list of unusual glycosylated molecules, which may be related to their ability to withstand a variety of stresses. Second, bacteria produce a wide range of glycan-binding proteins. These include adhesins that facilitate bacterial colonization, exotoxins that bind to host membrane glycans, and single-sugar-binding proteins involved in metabolism (see Chapter 42). Third, bacterial lifestyles are often defined by their capacity to digest and/or metabolize glycans. Complex nanomachines have been described for the degradation of complex carbohydrate structures. Assembling the various enzymes into a highly organized complex affords synergy and efficiency. For example, the roles of glycosyl hydrolases in biomass conversion are well documented (Chapter 59). The cellulose degradation machinery in Clostridioides sp. and other organisms comprises a suite of glycosidases and carbohydrate-binding modules, all assembled into a single surface complex (the cellulosome) via noncatalytic cohesin and dockerin structural modules. Within the gut microbiome, Bacteroidetes encode a remarkable array of carbohydrate-active enzymes in polysaccharide utilization loci, allowing them to adapt to dietary changes by sensing, binding, and degrading different complex substrates, followed by import of the products for catabolism. Indeed, bacteria within the many biomes of the human body derive benefit from foraging, not only on plant polysaccharides, but also on host-derived mucosal glycans. We continue to learn more about the intricate pathways for the digestion, capture, and catabolism of environmental and host carbohydrates.
ACKNOWLEDGMENTS
The authors acknowledge contributions to previous versions of this chapter by Jeffrey D. Esko, Tamara L. Doering, and the late Christian Raetz and appreciate helpful comments and suggestions from Graham Heberling and Jerry Eichler.
FURTHER READING
- Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71: 635–700. doi: 10.1146/annurev.biochem.71.110601.135414 [PMC free article: PMC2569852] [PubMed: 12045108] [CrossRef]
- Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol 8: 765–778. doi:10.1038/nrmicro2383 [PubMed: 20948550] [CrossRef]
- Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2: 1–16. [PMC free article: PMC2857177] [PubMed: 20452953]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11: 497–504. doi:10.1038/nrmicro3050 [PubMed: 23748339] [CrossRef]
- Nothaft H, Szymanski CM. 2013. Bacterial protein N-glycosylation: new perspectives and applications. J Biol Chem 288: 6912–6920. doi:10.1074/jbc.r112.417857 [PMC free article: PMC3591601] [PubMed: 23329827] [CrossRef]
- Angala SK, Belardinelli JM, Huc-Claustre E, Wheat WH, Jackson M. 2014. The cell envelope glycoconjugates of Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol 49: 361–399. doi:10.3109/10409238.2014.925420 [PMC free article: PMC4436706] [PubMed: 24915502] [CrossRef]
- Percy MG, Gründling A. 2014. Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu Rev Microbiol 68: 81–100. doi:10.1146/annurev-micro-091213-112949 [PubMed: 24819367] [CrossRef]
- Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83: 99–128. doi:10.1146/annurev-biochem-060713-035600 [PubMed: 24580642] [CrossRef]
- Artzi L, Bayer EA, Moraïs S. 2017. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol 15: 83–95. doi:10.1038/nrmicro.2016.164 [PubMed: 27941816] [CrossRef]
- Bontemps-Gallo S, Bohin J-P, Lacroix J-M. 2017. Osmoregulated periplasmic glucans. EcoSal Plus 7: doi:10.1128/ecosalplus.esp-0001-2017 [PubMed: 28593831] [CrossRef]
- Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2017. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14: 337–345. doi:10.1038/nrmicro.2016.25 [PMC free article: PMC4937791] [PubMed: 27026255] [CrossRef]
- Low KE, Howell PL. 2018. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr Opin Struct Biol 53: 32–44. doi:10.1016/j.sbi.2018.05.001 [PubMed: 29843050] [CrossRef]
- Mostowy RJ, Holt KE. 2018. Diversity-generating machines: genetics of bacterial sugar-coating. Trends Microbiol 26: 1008–1021. doi:10.1016/j.tim.2018.06.006 [PMC free article: PMC6249986] [PubMed: 30037568] [CrossRef]
- Powers MJ, Trent MS. 2018. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Molecular Microbiology 107: 47–56. doi:10.1111/mmi.13872 [PMC free article: PMC5740007] [PubMed: 29114953] [CrossRef]
- Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17: 403–416. doi:10.1038/s41579-019-0201-x [PMC free article: PMC6913091] [PubMed: 31142822] [CrossRef]
- Dulberger CL, Rubin EJ, Boutte CC. 2020. The mycobacterial cell envelope—a moving target. Nat Microbiol 18: 47–59. doi:10.1038/s41579-019-0273-7 [PubMed: 31728063] [CrossRef]
- Whitfield C, Wear SS, Sande C. 2020. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu Rev Microbiol 74: 1–23. doi:10.1146/annurev-micro-011420-075607 [PubMed: 32680453] [CrossRef]
- Whitfield C, Williams DM, Kelly S. 2020. Lipopolysaccharide O-antigens—bacterial glycans made to measure. J Biol Chem 295: 10593–10609. doi:10.1074/jbc.rev120.009402 [PMC free article: PMC7397119] [PubMed: 32424042] [CrossRef]
- AN OVERVIEW OF CELL ENVELOPE ARCHITECTURES
- PEPTIDOGLYCAN, A DYNAMIC STRESS-BEARING LAYER
- GRAM-POSITIVE BACTERIA PRODUCE ADDITIONAL CELL WALL GLYCOPOLYMERS
- MYCOBACTERIA POSSESS UNUSUALLY COMPLEX CELL ENVELOPE GLYCOCONJUGATES
- LIPOPOLYSACCHARIDE (ENDOTOXIN)—A KEY COMPONENT IN MOST GRAM-NEGATIVE BACTERIA
- PROTEIN GLYCOSYLATION—AN EXPANDING FACET OF BACTERIAL GLYCOBIOLOGY
- OSMOREGULATED PERIPLASMIC GLUCANS
- EXTRACELLULAR POLYSACCHARIDES
- ADDITIONAL FACETS OF BACTERIAL GLYCOBIOLOGY
- ACKNOWLEDGMENTS
- FURTHER READING
- Eubacteria - Essentials of GlycobiologyEubacteria - Essentials of Glycobiology
Your browsing activity is empty.
Activity recording is turned off.
See more...
