Open Access: This content is Open Access under the Creative Commons license CC-BY-NC-ND.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Mattick J, Amaral P. RNA, the Epicenter of Genetic Information: A new understanding of molecular biology. Abingdon (UK): CRC Press; 2022 Sep 20. doi: 10.1201/9781003109242-8

RNA, the Epicenter of Genetic Information: A new understanding of molecular biology.
Show detailsThe increasing sophistication of biochemical techniques developed between the 1960s and 1980s identified other relatively abundant RNA species beyond the canonical trio of tRNA, mRNA and rRNA. These included spliceosomal RNAs (snRNAs) that guide splicing and other aspects of gene expression; small nucleolar RNAs (snoRNAs) that guide modifications of rRNAs, tRNAs and snRNAs, over 700 of which are encoded in the human genome and are precursors of other types of small RNAs; 7SL RNAs, essential components of the ‘signal recognition particle’ that target proteins to the endoplasmic reticulum for membrane insertion or secretion; 7SK RNAs, which act as negative regulators of transcription; enigmatic Y RNAs involved in DNA replication and stress responses; ‘vault’ RNAs involved in recycling of cellular components in lysosomes and the plasticity of neuronal synapses; ‘virus-associated’ RNAs that modulate the innate immune response; and rodent brain-specific transposon-derived RNAs that modulate behavior. In the 1980s RNAs were discovered to have self-splicing and cleavage activities and to catalyze both translation and splicing, leading to the conclusion that RNA was the primordial molecule of life, subsequently outsourcing enzymatic functions to the more chemically versatile proteins and its information functions to the more stable and easily replicable DNA. These early observations and examples of the structural and functional versatility of RNA were, however, largely interpreted as relics rather than a new dimension, and failed to disturb the status quo.
The biochemical analyses of RNA in the 1960s detected many short RNAs in the nucleus and cytoplasm of eukaryotic cells a using new techniques of radioactive labeling, differential sedimentation and gel electrophoresis, and better procedures for isolating intact RNAs with detergents and chaotropic agents 1 to overcome degradation by RNases.
Initially there were concerns that the small RNAs are by-products of the biogenesis or degradation of larger RNAs. On the other hand, contrary to the generalization that nuclear RNAs are transient, destined for processing and export to the cytoplasm, the few groups studying these newly identified low molecular weight RNAs reported that they are highly expressed and account for ~20% of the nuclear RNA in mammalian cells. 2 They were also found to differ in size and sequence composition from tRNAs and rRNAs, and to be metabolically stable. 3
At least 10 discrete RNA species were identified, some of which contained methylated nucleotides, localized in specific subnuclear fractions (the nucleoplasm, chromatin or the nucleolus), with others in the cytoplasm. 4–9 Many of those RNAs in the nucleus were uridine-rich, leading to their designation as ‘U RNAs’, numbered in the order of their discovery as U1, U2, U3 and so on. 2 , 6 Robert Weinberg and Sheldon Penman named them “small nuclear RNAs” (snRNAs). 6 , 10
It was also found that RNA polymerase III is responsible for the transcription of many of these small RNAs, not RNA polymerase II, which transcribes mRNAs (and long non-protein-coding RNAs, Chapter 13), indicating that different RNA polymerases synthesize different classes of RNA. 11–13 RNA polymerase III products also include RNAs originating from repetitive sequences, only later characterized, such as those transcribed in human cells from Alu elements. 13 , 14
The characterization of snRNAs in the following decades revealed that they had ‘housekeeping’ functions in the modification and maturation of rRNAs, tRNAs and mRNAs, as well as other functions in gene regulation and cellular processes such as protein export. These years also saw RNAs encroach on the traditional domain of proteins, catalysis, which in turn led to a plausible explanation of the molecular origin of genetic information, with RNA at its core.
Spliceosomal RNAs
Although their roles were unknown, snRNAs were too small to function as mRNAs and, being mainly nuclear, did not seem to be directly involved in protein synthesis. On the other hand, some snRNAs contained sequences complementary to hnRNAs. This led Michael Lerner and Joan Steitz, and independently John Rogers and Randolph Wall, to propose in 1980 that these RNAs play a role in RNA splicing, 15–17 based on earlier work on sense-antisense interactions between mRNA and rRNAs sequences in translation initiation. 18 , 19 In particular, the complementarity of the U1 snRNA sequence to both the 5′ and 3′ splice site sequences of hnRNAs led to the hypothesis that splice sites are recognized and aligned through RNA–RNA interactions between the splice sites and U1 snRNA. 15–17
The characterization of the functions of these RNAs in the 1980s was aided by the fortuitous discovery that antibodies in the serum of individuals suffering autoimmune disorders, such as lupus erythematosus, precipitated ribonucleoprotein complexes (RNPs) containing snRNAs. 15 , 20–23 Several of the snRNAs interacted with a common antigen, the Sm (Smith) antigen, b named after the first lupus patient in whom such antibodies were detected. 20 , 23 Other autoantigens were associated with other RNP complexes. 25
These antibodies were used not only to purify the complexes but also to block the function of the corresponding snRNPs in vitro, which showed that splicing of pre-mRNA is inhibited by targeting the U1 RNP and therefore that snRNAs are required. 26 , 27 Chemical cross-linking confirmed that U1 and U2 RNAs do, in fact, base pair to hnRNAs in the nucleus, 28 , 29 and genetic complementation experiments confirmed that U1, U2, U4, U5 and U6 snRNAs function in splicing. 25 , 30 Characterization of the process revealed that large RNPs, which came to be known as ‘spliceosomes’, incorporate these snRNAs, wherein they interact with each other and with target pre-mRNAs to guide the process of splicing (Figure 8.1). 25 , 30–33

Figure 8.1
The mechanism of splicing and the complexity of the RNA interactions and structures (a) The RNA interaction network before the first trans-esterification reaction. The dotted lines indicate the triplex interactions at the catalytic core. (b) Three-dimensional (more...)
Much later, it was found that U1 and U4 snRNAs also regulate transcriptional initiation, transcript structure and chromatin architecture, 34–40 and that U2 snRNA is required for RNA polymerase II pausing, 41 coupling transcription to splicing, which are intertwined processes (Chapters 14 and 16). It was also discovered that there is a minor class of spliceosome, which contains specific snRNAs (U11, U12, U4atac and U6atac) equivalent to but distinct from their counterparts in the major U2-type spliceosome (U1, U2, U4 and U6), and which recognizes a rare class of introns initially referred to as AT–AC introns, now called U12-type introns. 42–45 The minor spliceosome is required for development and may have particular functions in the brain. 45–48
Small Nucleolar RNAs
Other small RNAs had other functions. The highly conserved U3 RNA was found to associate with 28S rRNA 8 , 49 and to be localized in the nucleolus, 11 , 50 where ribosome biogenesis occurs, which led Jean-Pierre Bachellerie to suggest in 1983 that U3 and other “small nucleolar RNAs” (snoRNAs) c participate in this process. 50 Subsequently it was also found that autoimmune antibodies recognizing a nucleolar protein, fibrillarin, 53 would co-precipitate not only U3 but also the less abundant U8 and U13 RNAs. 54 It was later found also to bind U16. 55
Similar to the observations that led to the elucidation of the roles of snRNAs in pre-mRNA splicing, snoRNAs were found to have short sequence motifs complementary to rRNA sequences, which indicated that snoRNAs were involved via base pairing in rRNA processing, modification or other aspects of ribosome biogenesis. 50
Nevertheless, it was only in 1990 that U3 was shown to be essential for rRNA processing, 56 and in 1996 that snoRNA U24 directs site-specific methylation of rRNAs 57 with subsequent studies showing that other snoRNAs perform similar functions via base pairing with target sequences adjacent to modification sites. 51 , 58 Thus, although first identified in the 1960s, it was three decades before snoRNAs were defined as a new “class of RNAs” with demonstrated functions. 51 , 59
SnoRNAs are ~60–300nt in length and are classified into two families (based on typical sequence motifs and structural features) that guide enzyme complexes to perform 2′-O-ribose methylation (‘C/D box’ RNAs) or pseudouridylation (‘H/ACA box’ RNAs) respectively of target nucleotides (Figure 8.2), not only in rRNAs and tRNAs but also in snRNAs. 60 Homologs of C/D box and H/ACA box snoRNAs occur in archaea, where they also guide modifications of tRNAs, indicating that they first evolved over 3 billion years ago. 61–63
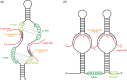
Figure 8.2
Schematic representation of C/D (a) and H/ACA (b) box snoRNAs. Consensus box sequences are highlighted in green. The conserved structures within these snoRNAs guide effector protein complexes that catalyze 2′-O-methylation or pseudouridylation (more...)
There are also many snoRNAs that show tissue-specific expression and whose targets are unknown, described as “orphan” snoRNAs. 60 Some were later found also to be involved in RNA processing, including the C/D box snoRNAs U8, U14, and U22, as well as H/ACA box snoRNAs snR10, snR30, E2 and E3, which direct site-specific cleavage of pre-rRNAs. 65 Yet, other snoRNAs were shown to regulate alternative splicing by base pair recognition 66 and to guide other modifications such as acetylation of specific cytosine residues in 18S rRNA. 67
More was to come. In 2001, Beáta Jády and Tamás Kiss identified a sno-like RNA, U85, containing both H/ACA and C/D box motifs, which is localized in ‘Cajal bodies’ (a subnuclear domain associated with nucleoli, discovered by Santiago Ramón y Cajal in 1903 68 ) and guides 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal snRNA. 69 The snRNA U7 is also localized in Cajal bodies and participates in histone pre-mRNA 3′ end formation. 70 , 71 Other small Cajal body-specific (‘sca’) RNAs have since been identified, some with a composite structure similar to U85, while others have only H/ACA box or C/D box domains, which guide modifications of spliceosomal snRNAs 72 and tRNAs, the latter as part of a stress response. 73 , 74 The RNA component of the human telomerase complex also contains a characteristic H/ACA box scaRNA-like structure and also localizes in Cajal bodies. 75–77 Primate-specific H/ACA snoRNA-like RNAs, called AluACA RNAs, are derived from intronic Alu-repeat RNAs 78 and new functions of snoRNAs continue to be discovered, such as the maintenance of chromatin accessibility. 79
There are over 700 known snoRNA and snoRNA-like RNAs encoded in the human genome. 80 Most snoRNAs are produced by processing of intronic RNAs excised from host transcripts, 81 commonly those of genes encoding proteins involved in translation or ribosome biogenesis, including ribosomal proteins, translation factors and nucleolar proteins such as fibrillarin, 60 , 82 the first evidence of parallel genetic output. Many other snoRNAs are derived from the introns of transcripts that do not encode proteins, 83–88 some involving species-specific alternative splicing, 89 and whose primary function in many cases is uncertain, although others, like Gas5, 90 have demonstrated roles as long regulatory RNAs (Chapters 9 and 13). Some snoRNAs are expressed exclusively in the brain. 51 , 91–93
The complexity of the relationships between small RNAs is illustrated by the later discovery that snoRNAs, from yeast to humans, are processed to produce three subspecies, one of which functions as a microRNA in the RNA interference pathway 64 , 94–96 (Chapter 12). The complexity of the networks, and an indication of how much is yet to be understood about them, is highlighted, for instance, by the observation that a human-specific snoRNA is attached to the end of a longer non-coding RNA that regulates rRNA biogenesis and nucleolar structure (via phase separation, Chapter 16). 97–99 In addition, many snoRNAs accumulate in the form of stable lariats instead of fully processed snoRNP particles, with as yet unknown functions. 100
Aberrant expression of snoRNAs has been linked to human disease. There is a large cluster of C/D box snoRNAs in a parentally imprinted gene that is normally expressed in the brain from the maternally derived allele, perturbations of which are associated with Angelman and Prader-Willi syndromes. 88 , 91 , 101 , 102 One of the snoRNAs in this region, HBII-52, contains an 18 nucleotide sequence that is complementary to an exon in the serotonin receptor 5-HT(2C) mRNA and mediates its alternative splicing. 103
Other Small Guide, Scaffolding and Regulatory RNAs
7SL and 7SK RNA species, their names reflecting their sedimentation coefficient, were identified by Penman in 1976. 104 These RNAs were initially thought to have a viral origin 3 but were shown to be present in uninfected cells. 104 , 105
7SL RNA is ~300nt in length. It is ubiquitous in eukaryotic cells and was found, accidently, d to be an essential component of the protein export ‘signal recognition particle’ (SRP), 106 characterized in the 1970s and 1980s by Günter Blobel and colleagues. 107 The SRP associates with the ribosome and targets nascent proteins to the endoplasmic reticulum via an N-terminal ‘signal’ or ‘leader’ sequence for membrane insertion or secretion into the extracellular milieu, 106 , 108 , 109 with 7SL RNA acting as a scaffold upon which the six proteins of the SRP assemble. 109 , 110 Similar RNAs were later shown to be involved in protein export in bacteria and archaea. 111 , 112
7SL RNA was subsequently found to be required for the selective packaging of the RNA/DNA modifying enzymes APOBEC3G and 3F into retroviral particles 113–117 and to repress the translation of the tumor suppressor TP53. 118 It is also a precursor of the Alu elements in the human genome (Figure 8.3). 119–121

Figure 8.3
The structure of 7SL RNA, showing the part coopted into Alu transposable elements in primates. SPR19 binding sites are shown. (Reproduced with minor modifications from Itano et al. with permission from John Wiley and Sons.)
7SK RNA is highly expressed in vertebrates and, like most RNAs, has a complex structure. 122 Early evidence indicated that 7SK regulates transcription and transcription termination in a tissue- and species-specific manner, 123 , 124 but mechanistic insights would not emerge until the early 2000s. These studies showed that 7SK RNA acts as a trans-acting negative regulator, uncovered serendipitously in biochemical assays to identify factors that regulate RNA polymerase II, similar to the “general transcription factor” role identified for the small 6S RNA in bacteria 125 (Chapter 9), but having additional roles in gene expression regulation in animals 126–134 (Chapter 13). 7SK is also present in invertebrates, and, despite the fact that it is little primary sequence similarity, its identification was possible due to its conserved secondary structural motifs and domains. 135 , 136
In one of the earliest demonstrated regulatory RNA roles, in 1980, Hugh Pelham, Robert Roeder and colleagues discovered that the transcription factor TFIIIA e not only binds the 5S rRNA gene f but also its transcript, which results in a feedback loop that titrates the transcription factor away from the gene, inhibiting further transcription and stabilizing the transcript until required for ribosome assembly. 140 , 141
Y RNAs were identified in 1981 as components, like snRNAs, of autoantigens in systemic lupus patients. 21 , 142 There are four distinct and highly conserved Y RNAs (in humans ranging from ~80 to ~110nt), 143 which are structural components of the Ro autoantigen. 144–146 The Ro protein, lack of which causes a lupus-like syndrome in mice, appears to prevent autoimmunity by recognizing misfolded RNAs, with Y RNAs regulating the process. 147–149 Y RNAs also occur in bacteria where they associate with orthologs of mammalian Ro and are involved in rRNA maturation and stress responses, 150–152 with a modular structure that includes a domain that mimics tRNAs, 153 indicating an ancient function in cellular RNA biology.
Vault RNAs, short 80–150nt RNAs transcribed by RNA polymerase III, so named because of their presence in large ovoid ribonucleoprotein particles in the cytoplasm of eukaryotic cells that resemble the arches of cathedral vaults, were discovered in 1986 by Nancy Kedersha and Leonard Rome. 154 , 155 Vault RNAs are considered essential for eukaryotic cell biology because of their high conservation and near ubiquitous presence. 156 Their function is not well understood, but recent evidence indicates that they play a role in regulating autophagy, 157 i.e., the degradation and recycling of cellular components in lysosomes, 158 as well as apoptosis 159 , 160 and signaling pathways involved in neuronal synapse formation and plasticity. 161
Other RNAs discovered in these years were the highly abundant viral RNAs in infected cells, some of which interact with autoantigens. The ~160nt VA RNAs (virus-associated RNAs) present in cells infected with adenovirus 163 (Figure 8.4) were in fact the first non-coding RNAs described after the ‘canonical’ RNAs (rRNA, tRNA and mRNA) and the first non-coding RNAs shown to be expressed from mammalian viruses, reported in the same year as the identification of the lac repressor. VA RNAs were later shown (among other roles) to inhibit protein kinase R (PKR) g to curb the innate immune response and enhance the translation of viral RNAs. 163–169
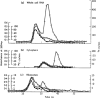
Figure 8.4
The discovery of low molecular weight virus-associated RNA (VA RNA): size chromatography of newly synthesized (radiolabeled) RNA isolated from infected (open circles) and uninfected (closed circles) cells infected with adenovirus (UV absorbance shown (more...)
RNAs that are not highly conserved across large evolutionary distances but have specific expression patterns were also identified during the 1980s. Examples include neuronal RNAs that are transported into dendrites, such as B2, a ~180nt RNA transcribed by RNA polymerase III from repeated sequences in mice, which shows higher expression in some tumor cells and heat-shocked cells, 170–172 and BC1 (brain cytoplasmic RNA 1), a ~150nt RNA transcribed in rats from repeated sequences derived from a tRNA, 173–175 with a human equivalent, BC200. 176 , 177 Transgenic mice lacking BC1 have no obvious developmental deficiency, but display reduced exploration activity and increased anxiety, a phenotype that is invisible in the cage but likely lethal in the wild. 178
Small RNAs are also required as primers for DNA replication 179 , 180 and for the maintenance of telomeres, h shown by Elizabeth Blackburn, Carol Greider and Jack Szostak to be accomplished by retrotransposon-derived RNAs operating on repeat sequences to replicate chromosome ends. 182–189
Small common RNAs such as snRNAs and snoRNAs were relatively easy to find. Others were identified later in genomic datasets by characteristic sequence motifs and secondary structures, using a growing suite of bioinformatic tools 190–193 and databases such as Rfam 194 , 195 and RNAcentral. 196 On the other hand, lower copy number transcripts that are less conserved and expressed only in particular circumstances or cells were difficult to detect, a problem compounded by the lack of anticipation of the existence of cell-specific transcripts beyond mRNAs and their nuclear precursors (Chapters 12 and 13).
Catalytic RNAs and the Ancient RNA World Hypothesis
The participation of RNAs in a variety of cellular processes and the existence of many RNPs with different functions signaled that RNAs are versatile. As put by Crick in 1966, referring to the ability of RNA to form complex secondary structures: “tRNA looks like Nature’s attempt to make RNA do the job of a protein.” 197
In 1957, an influential symposium in Moscow on the origin of life speculated that RNA most likely preceded proteins in the origin of life, a view supported there by Brachet, Mirsky, Oparin i and others. 199 In 1962, Alex Rich proposed that RNAs had a central role in the origin of life. 200 In the late 1960s, Orgel, Woese and Crick also hypothesized that RNAs might have preceded proteins in a pre-cellular world, predicting that RNAs possessed the required enzymatic activities. 197 , 201 , 202
Nonetheless, the existence of catalytic RNAs in extant organisms was completely unexpected. 203 In 1982, Tom Cech and colleagues discovered that RNA can perform autocatalytic ‘self-splicing’ rearrangements, removing the intervening sequence of rRNA precursors by excision and cyclization in the ciliate protozoan Tetrahymena. They named these catalytic RNAs “ribozymes”, 204 later categorized as ‘self-splicing group I introns’.
Group II self-splicing introns were recognized in 1983 in organelle genomes by François Michel and Bernard Dujon. 205 They consist of a catalytically active intron RNA and an intron-encoded reverse transcriptase, enabling intron proliferation within genomes. Group II intron RNA catalyzes its own splicing via transesterification reactions that are the same as those of spliceosomal introns, yielding spliced exons and an excised intron lariat RNA. 206–208 Thus, group II introns appear to be the ancestors of modern spliceosomal introns 209–213 and likely entered the eukaryotic lineage through the bacterial ancestor of the mitochondrion. 214 , 215
Also in 1983, Sidney Altman, Norman Pace and colleagues showed that RNA is the catalytic component of the bacterial RNase P complex, which produces mature tRNAs by cleaving a 5′ end sequence in a process analogous to splicing. 216 The RNA in RNaseP is one of the only two ribozymes found in all domains of life: 217 a closely related eukaryotic RNA was also described in the early 1980s 218 and later shown to be the catalytic center of a complex involved in sequence-specific processing of mitochondrial and other RNAs, 219 hence called RMRP (RNase MRP), although it is mainly located in the nucleolus and has other important regulatory functions (Chapter 13) (Figure 8.5). 220 , 221
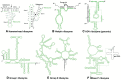
Figure 8.5
Secondary structures of six types of ribozymes. (Reproduced from Takagi et al. with permission of Oxford University Press. The ribozyme or intron portion is printed in green. The substrate or exon portion is printed in black.)
The demonstration that RNA molecules are able to cleave and join themselves or other RNAs, and capable of the phosphodiester bond transfers needed for RNA synthesis, prompted Walter Gilbert to formalize the “RNA World” hypothesis. 223 In this view, RNA molecules, not proteins, were the precursors of existing life, having performed the catalytic and information storage functions j in the pre-cellular world. k
Since then self-cleaving ribozymes have been found in bacteria, protists, fungi, plants, nematodes, arthropods, insects and vertebrates, including human, 222 , 227–234 one of which has been shown to methylate other RNAs. 235 More surprisingly for its implications, B2 and Alu elements – ‘repetitive’ sequences that occur in ~350,000 copies and over 1 million copies in the mouse and human genomes respectively – have been shown to harbor self-cleaving ribozyme activity that is induced upon stress and T-cell activation by binding to the Polycomb histone methyltransferase protein EZH2 233 (Chapter 16).
It has been reasonably postulated that modified nucleotides may have enhanced the early catalytic capacities of RNAs and facilitated their path to self-replication. 236 Ribosomal RNAs, small nucleolar RNAs and spliceosomal RNAs are all heavily modified, and RNA modification has been widely deployed as a mechanism to introduce plasticity into RNA regulatory circuits (Chapter 17).
The Catalytic Heart of Splicing and Translation
In 1992, Harry Noller and colleagues showed that rRNA is not just a structural scaffold for the ribosome, as had been widely assumed, but harbors the central peptidyl transferase activity for protein chain extension in translation, making the ribosome a complex and conserved ribozyme 237 , 238 (Figure 8.6). RNAs must therefore have pre-existed proteins.

Figure 8.6
The active site of the ribosome, the peptidyl transferase center, is located on the large ribosomal subunit within a highly conserved region of the ribosomal RNA (red). (a) Model of RNA structure. (b) detail. (Image courtesy of Marina Rodnina and Wolfgang (more...)
As noted above, RNA splicing is also an RNA catalyzed reaction, l with sequence and mechanistic similarities to group II self-splicing introns in bacteria, capable of inserting into new locations by reversal of the splicing reaction. 209 , 239
Many small molecules, including antibiotics, target ribosomal and other RNAs: these include tetracyclins, aminoglycosides such as streptomycin and gentamycin, chloramphenicol, carbomycin A, blasticidin S, puromycin and hygromycin B. 240–242 They also include poisons such as ricin, an N-glycosidase RNA-modifying enzyme that depurinates a conserved loop of 28S rRNA and leads to irreversible arrest of protein synthesis. 243 Indeed, therapeutic targeting of diverse RNA types by small molecules is an area of rapidly growing interest.
The Digital and Analog Faces of RNA
This period also saw the expansion of RNA structural biology, led by Noller, Eric Westhof, Tom Steitz, Robin Gutell, Jennifer Doudna and others, who showed that RNAs have extraordinarily complex structures, capable of binding proteins, as a consequence of being able to form hydrogen bonds on all three faces – the Watson-Crick face, the Hoogsteen face (within the double-stranded groove) and the ribose, because of its 2′ hydroxyl, which is lacking in DNA. 244–246 They also have exposed sequences that can base pair with other RNAs and DNA, through RNA:RNA duplexes, R-loops (RNA:DNA duplexes with displaced single stranded DNA) and RNA:DNA:DNA triplexes, which are common in eukaryotic chromatin 247 , 248 (Chapter 16).
The structural versatility of RNA has been explored and exploited in vitro. In the 1990s, the groups of Larry Gold and Jack Szostak developed SELEX (‘Systematic Evolution of Ligands by Exponential Enrichment’) to evolve artificial RNAs that bind specific ligands (‘aptamers’) or have other activities, 249–252 speculating that the same will have occurred in vivo. 253
In addition, “free” low molecular weight circular RNA molecules lacking protein-coding capacity but having “peculiar” secondary structures and ribozyme activity were discovered in the 1970s to infect and autonomously replicate in plant cells (baptized as ‘viroids’), 254 , 255 and postulated to represent living fossils 256 subject to Darwinian evolution in a prebiotic world. 257 Other curious virally associated RNAs were also described in the late 1970s and 1980s. 258–260
Candles in the Dark
By 1985, it was becoming apparent that RNAs are multifaceted molecules that have specific subcellular locations, form complex structures, interact with (many) proteins and perform a vast array of functions beyond protein synthesis, from gene regulation to acting as components of cellular complexes, catalytic molecules and antisense guides. These observations did little to challenge the assumption that the destination of genetic information is (nearly always) the production of a protein and were regarded as interesting but idiosyncratic additions to the tapestry of molecular biology, rather than the first indications of a wider role for RNA in cell and developmental biology.
In his 1986 article describing the RNA World hypothesis, Gilbert proposed that, after the emergence of DNA as the carrier of genetic information, RNA was then “relegated to the intermediate role that it has today – no longer the centre of the stage, displaced by DNA and the more effective protein enzymes”. 223 Others concurred. 261
Penman lamented in 1991:
If genes just make proteins and our proteins are the same, then why are we so different? … we have the bizarre proposal dominating biology that the incredibly complex living systems are described entirely by component proteins and their coding sequences. Where is the genetic information that executes the design of an organism? We do not have to look far for a candidate. There is plenty of information in the more than 95% of the genome that is devoid of open reading frames. These sequences, heavily transcribed in all cells, appear to have little, if anything, to do with making proteins. 262
Further Reading
- Darnell J.E. (2011) RNA: Life’s Indispensable Molecule (Cold Spring Harbor Laboratory Press, New York).
- Gesteland R.F., Cech T. and Atkins J.F. (2006) The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World (Cold Spring Harbor Laboratory Press, New York).
- Joyce G.F. (1991) The rise and fall of the RNA world. The New Biologist 3: 399–407. [PubMed: 1712228]
- Lazcano A. (2012) The biochemical roots of the RNA world: From zymonucleic acid to ribozymes. History & Philosophy of the Life Sciences 34: 407–423. [PubMed: 23316569]
- Mount S.M. and Wolin S.L. (2015) Recognizing the 35th anniversary of the proposal that snRNPs are involved in splicing. Molecular Biology of the Cell 26: 3557–3560. [PMC free article: PMC4603926] [PubMed: 26463979]
- Müller, F., et al. (2022) A prebiotically plausible scenario of an RNA–peptide world. Nature 605: 279-284. [PMC free article: PMC9095488] [PubMed: 35546190]
- Robertson M.P. and Joyce G.F. (2012) The origins of the RNA world. Cold Spring Harbor Perspectives in Biology 4: a003608. [PMC free article: PMC3331698] [PubMed: 20739415]
- Vicens Q. and Kieft J.S. (2022) Thoughts on how to think (and talk) about RNA structure. Proceedings of the National Academy of Sciences USA 119: e2112677119. [PMC free article: PMC9169933] [PubMed: 35439059]
- Wilkinson M.E., Charenton C. and Nagai K. (2020) RNA splicing by the spliceosome. Annual Review of Biochemistry 89: 359–388. [PubMed: 31794245]
Footnotes
- a
Also in bacteria (Chapter 9).
- b
Sm proteins participate in a wide variety of RNA transactions and predate the Archaea-Eukarya split. 24
- c
- d
Blobel and colleagues were expecting only proteins to be components of the SRP. The presence of RNA was revealed by a strong UV absorbance signal at a wavelength typical of nucleic acids in the SRP preparation, detected as a result of an incorrect setting on the detector. 106
- e
TFIIIA was shown by Aaron Klug and colleagues to interact with RNA and DNA via repetitive domains stabilized by zinc, called ‘zinc fingers’. 137 Zinc finger transcription factors were later found to be the largest class of transcription factors in plants and animals, comprising 3% of the genes in the human genome 138 (Chapter 16).
- f
A non-coding RNA, termed 5S-OT, is transcribed from 5S rDNA loci in eukaryotes and has been shown to regulate transcription of 5S rRNA in mammals. An antisense Alu element has inserted at the 5S-OT locus in monkeys, apes and humans and regulates alternative splicing of other genes via Alu/anti-Alu pairing. 139
- g
PKR is in fact variously inhibited or activated by trans-acting RNAs. 162
- h
The existence of telomeres to protect chromosome ends was inferred by McClintock and Muller in the 1930s. Muller coined the term ‘telomere’ from the Greek ‘telos’ (end) and ‘meros’ (part). 181
- i
Oparin previously had a long running debate with Hermann Muller, with Oparin maintaining that life was the outcome of a step-wise process of pre-cellular evolution of membrane-bound polymolecular systems, whereas Muller argued that life started with the appearance of the first nucleic acid molecule. 198
- j
This view is supported by many observations, including that an RNA polymerase ribozyme obtained by in vitro evolution can copy complex RNA templates, including itself, albeit at low fidelity. 224 There are also plausible scenarios for the prebiotic synthesis of the pyrimidine and purine building blocks of RNA. 225
- k
RNA can also nucleate ‘liquid crystal’ phase-separated domains, explored early on by Oparin (Chapter 2), who worked extensively on the role of RNA as a polyanion in the formation of ‘coacervates’, 226 a property that would come to the fore as central to both modern cell biology and prebiotic evolution as RNAs would have been able to sequester organic molecules in a proto-cell (Chapter 16).
- l
U2 and U6 snRNAs interact to form the conserved structure of the catalytic triplex, coordinating two magnesium ions to form the active site of the spliceosome. 212
- The Expanding Repertoire of RNA - RNA, the Epicenter of Genetic InformationThe Expanding Repertoire of RNA - RNA, the Epicenter of Genetic Information
Your browsing activity is empty.
Activity recording is turned off.
See more...