NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.
This publication is provided for historical reference only and the information may be out of date.
Scope
This chapter provides an overview of the representation of sequence variation in NCBI’s databases and a summary of the tools that are available to access and use these data. The resource-specific chapters in this section provide all the details; this overview ties these chapters together and fills in gaps where chapters are not yet available. The variation home page (http://www.ncbi.nlm.nih.gov/variation) is NCBI’s portal to both databases and tools related to variation.
History
The major databases representing variation at NCBI are the databases archiving information about the location and types of variation, namely dbSNP for variation less than about 50 base pairs (bp), and dbVar for longer, structural variation. Those data are then made accessible from several sites at NCBI (e.g., Gene, Nucleotide, RefSeq) or have value added by establishing connections between variations and multiple types of phenotypes including disease names, clinical features, and gene expression (ClinVar, dbGaP, and PheGenI). Representation of variation at NCBI includes all taxa for which submissions have been received. This ranges from viruses to bacterial pathogens to human. The variation does not have to be heritable; sequence variation that has been observed in tumors or other somatic sources is also represented.
Although information about variation is maintained in distinct databases, the representation in those databases is being standardized to improve searching, reporting, evaluation, and analysis. For example, representation of types of variation (single nucleotide, insertion, copy number gain), of consequences of that variation (nonsense, missense, frameshift), and functional consequences (exon loss) are harmonized to terms from Sequence Ontology (http://sequenceontology.org/). With the launch of ClinVar in 2013, standardization reporting of clinical significance is being shifted from the archival databases to ClinVar.
dbSNP and short variation
A major focus of sequencing projects is to identify variations and evaluate their consequences. Beginning in 1998, the National Center for Biotechnology Information (NCBI) established a database, dbSNP (http://www.ncbi.nlm.nih.gov/snp), to manage information about human variation. Even then, the scope of the database was not limited to storing information about single nucleotide polymorphisms (SNPs), rather submission of all types of variation were accepted without restriction by allele frequency.
From the beginning, dbSNP has assigned an accession to each submitted variation (the submitted SNP or ss identifier). Multiple submissions for the same variation and their attributes are integrated to create a reference record (reference SNP or rs identifier. Allele frequency observed in particular populations is also accepted, but not required.
The overwhelming majority of early submissions to dbSNP was in support of HapMap (http://hapmap.ncbi.nlm.nih.gov) and represented single nucleotide variations that were indeed polymorphic according to the definition of a minor allele frequency of at least 1%. Thus there is a common misconception that if a variation were in dbSNP, it was indeed a polymorphic single nucleotide change.
dbSNP continues to support tools and reports geared to diverse users, from population geneticists to medical geneticists. An example is the NCBI Variation Viewer tool that has recently been completely rewritten to report all types of human variation.
dbVar and structural variation
dbVar (http://www.ncbi.nlm.nih.gov/dbvar) archives information about genomic structural variation from studies submitted for any organism. In general, these variants are longer than 50 bp. Each variant instance is assigned an identifier beginning with nssv. One or more variant instances at the same location are assigned an identifier beginning with nsv. This identifier marks a region of the genome that a submitter has defined as containing structural variation. Variant regions point to sets of exemplar variant instances which support the assertion that the region contains variation. Because dbVar exchanges data with DGVa (1), some records may have accessions beginning with essv or esv, for instances and regions respectively.
As the archival databases (dbSNP and dbVar) became established, more and more data were being generated to use that variation to improve our understanding of population genetics, identify regions of the genome that affect rare and common disorders, and identify the effect of variation on gene expression. Thus dbSNP, which originally archived all those data, began to spin off or collaborate with studies having specific scopes. These include the resources in the table below:
| HapMap | www | Human population structure; identification of blocks of linkage disequilibrium and common variation |
| 1000 Genomes | International project; dbSNP and dbVar maintain identifiers for locations where variation has been observed | Understanding of variation in more populations of apparently healthy individuals |
| Genotypes | No dedicated interface | Maintains the genotype information from 1000 Genomes and GO-ESP. Supports displays in our 1000 Genomes and Variation View browsers |
| PheGenI |
http://www | Interface to view associations of phenotype to common variation |
| ClinVar |
http://www | History of interpretation of medically important variation |
Data Model
Archive submissions
A major function of dbSNP and dbVar is to archive submissions. Thus each manages information about the submitter, the date of the submission, the study that generated the data, as well as the content. Part of the archival function includes validating the submission, for example determining if the data are consistent with the genome for which the submission was provided. These archives are accessioned, assigned ss identifiers by dbSNP and nssv by dbVar as appropriate.
ClinVar also archives submissions, namely the interpretation of sequence variation relative to health status. These submissions are assigned a 12-character accession beginning with the letters SCV and followed by numbers padded to 9 places. If submitters submit an update, the accession is assigned a new version.
Aggregate data
dbSNP aggregates data from multiple submissions by location on the genome and type of variation. The result of this aggregation is assigned a refSNP (rs) identifier, which is commonly used to reference that variant location in subsequent studies and publications. It must be emphasized that the rs identifier does not indicate the explicit sequence change at a location. In other words, one rs is assigned to a location on the genome where there is single nucleotide variation, even if all 4 nucleotides have been observed at that location.
ClinVar aggregates data based on the combination of the variation and phenotype. These aggregates are assigned a12-character accession beginning with the letters RCV and followed by numbers padded to 9 places. The RCV accessions are versioned, with a new version assigned if an SCV is updated or a new SCV is added to the set.
Interpret data
Variation resources do compute some interpretations of the archived information. For example, for human variants, dbSNP and dbVar determine the HGVS representations of variants (http://www.ncbi.nlm.nih.gov/variation/hgvs). dbVar and ClinVar compute ISCN coordinates based on sequence location; the variation group calculates the molecular consequences of a sequence change based on an NCBI Annotation Release. Other interpretations, such as clinical significance, functional consequence of variation, or association results are represented only as submitted.
Access
The variation resources provide interactive access via the Web, application-based access via E-Utilities or other API, data extractions for FTP transfer, and specialized downloads. The variation portal page is the recommended starting point to discover variation information of interest.
Related Tools
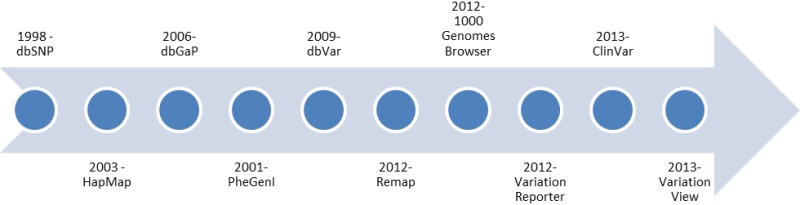
Figure 1.
Overview of the introduction of variation-related resources at NCBI.
Clinical Remap
The NCBI Clinical Remapping Service projects variant coordinates that are on a RefSeqGene or LRG to an assembly, or coordinates from an assembly to any available RefSeqGene or specified list of target RefSeqGenes or LRG.
Because the Clinical Remap Service accepts BED, GVF, HGVS and VCF formats, it can be used to view novel variations with asserted positions in a larger genomic context. When queried, the Clinical Remap service provides a full mapping report, a variation report that shows known dbSNP variants (including those with predicted consequences or with associated clinical information) that map to the same position, as well as an Annotation Data Report and Genome Workbench Files that you can use for further data analysis.
ClinVar
ClinVar is a database that archives the relationship between variations and their possible phenotypes by collecting variations found in patient samples, clinical assertions made about each variant, and the associated data supporting a particular clinical assertion. The ClinVar database can be used as a gateway to additional phenotypic information for a variant including:
- Condition(s) asserted to be associated with the variant
- Available evidence supporting a particular clinical assertion for the variant
- Current interpretation of clinical significance
Links from ClinVar Summary and Individual Accession Reports to related dbSNP records are located in the “Allele Description” section and the SNP track “Sequence View” section.
Although refSNP report pages provide an assertion of clinical significance when available, they currently do not link to ClinVar records as of this writing. dbSNP anticipates reciprocal links to ClinVar from rs report pages in the future.
dbGaP
dbGaP archives and distributes data from studies that examine the relationship between phenotype and genotype. Such studies include Genome-Wide Association Studies (GWAS), medical sequencing, and molecular diagnostic assays. dbGaP allows open access for non-sensitive data, including study documents, phenotypic variables, and genotype-phenotype analyses, but controlled access for restricted data that include pedigrees, pre-computed genotype/phenotype associations, as well as de-identified phenotypes and genotypes of study participants.
Links are available from dbGaP controlled-access records to related variation data in dbSNP, but there are no reciprocal links from dbSNP records to dbGaP since dbGaP data security measures prohibit access to dbGaP individual-level data from any external resource. The refSNP report “Association” section will link to association results from NHGRI_GWAS and/or PheGenI when association data is available.
dbMHC
dbMHC provides a platform where users can access, submit, and edit data related to the human Major Histocompatibility Complex, also called the HLA (Human Leukocyte Antigen).
Both dbMHC and dbSNP store the underlying variation data that define specific HLA alleles. dbMHC provides access to related dbSNP records at the haplotype and variation level, whereas dbSNP provides access to related dbMHC records at the haplotype level.
Gene
The Gene database is the NCBI resource for gene-related data. Gene provides a Variation section that provides links to variation data and tools when data are known to be available.
HapMap
The International HapMap Project site allows access to its catalog of statistically related variations, also known as haplotypes, for a number of different human populations, and is a useful resource for those researchers looking for variations associated with a particular gene. HapMap haplotypes can be searched by a landmark such as a refSNP number or gene symbol, as well as by sequence region or chromosome region. The resulting HapMap report includes an ideogram with various tracks that can be altered to provide required data, and appropriate tracks in the report will provide direct links to refSNP cluster records.
Phenotype-Genotype Integrator (PheGenI)
PheGenI allows an investigator to use clinical or physical traits, gene name, chromosomal location, or a dbSNP ID number (ss or rs) to search for, view, or download data from various NCBI resources in a single response page. A PheGenI search result can include the following data, if available:
- Related association results from Genome-Wide Association Studies (GWAS) that include links to associated PubMed abstracts
- Related dbGaP study data
- Related expression Quantitative Trait Loci (eQTL) data
- An annotated table of related genes that includes associated OMIM links
- An annotated table of variations from dbSNP
- An interactive view of the genome decorated with search results
PheGenI can be accessed from dbSNP using the PheGenI link, located in the refSNP report “association” field. This link will not be available if the record does not have related association data. PheGenI is also accessed from Gene, via a link provided at the top of the Phenotypes section, labeled Review eQTL and phenotype association data in this region using PheGenI.
PubMed
PubMed may provide access to information about variation that is not yet in a specific database. For example, this query http://www.ncbi.nlm.nih.gov/pubmed/?term=%28mutation[title]+OR+variation[title]%29+AND+novel[title] will retrieve articles with either mutation or variation in the title and novel in the title, and even be restricted to those for which free full text is available (http://www.ncbi.nlm.nih.gov/pubmed/?term=%28mutation[title]+OR+variation[title]%29+AND+novel[title]%20AND%20%22loattrfree%20full%20text%22[sb]).
Variation Batch Submission (VarBatch)
VarBatch is an online, spreadsheet-based interface to facilitate submitting and updating information about human variations described as HGVS expressions. When an asserted clinical variation is processed through VarBatch, it is assigned both a dbSNP submitted SNP (ss) accession as well as a ClinVar accession (format: SCV000000000.0), since the ClinVar accession represents the asserted variation/phenotype relationship.
Variation Reporter
Variation Reporter matches submitted variation calls to variants in NCBI’s databases, and reports back metadata that NCBI has for matching variants. If a variant is novel to NCBI, and the variation is near a feature annotated by NCBI, Variation Reporter will report the predicted molecular consequence based on changes to that annotated feature.
Variation Viewer
Variation Viewer allows a user to review variation in the context of multiple types of sequence features and filter results to a subset of interest. The user can select the assembly; search for features such as genes across the genome; upload local data to view in the context of what is available from NCBI; navigate by gene symbol, exon, rs#, nssv accession, cytogenetic band; retain a history; and filter displays by Variant type, Molecular consequence, minor allele frequency from 1000 genomes (1000 Genomes MAF), minor allele frequency from GO-ESP, and representation in dbSNP, dbVar, and ClinVar. The sequence display, based on NCBI’s graphical viewer, provides multiple track options including segmental duplications, paralogous regions, annotation releases from NCBI and Ensembl, somatic variants, common variants, RNAseq support of intronic features, and repeats. The track selection is designed to support evaluation of variations in the region of interest.
1000 Genomes Browser
The 1000 Genomes Browser provides access to 1000 Genomes data, including variations, genotypes, and sequence read alignments within the context of GRCh37, the reference assembly used by the 1000 Genomes Project (2) for analysis. The browser allows you to configure the display to include multiple data tracks of interest, and provides links to related data housed in various NCBI resources. The 1000 Genomes Browser allows users to quickly view variation, allele frequencies or counts by population group, and alignments of reads to support review of the evidence used in calling the sequence. Detailed instructions about how to use the browser are provided.
References
- 1.
- Lappalainen I, Lopez J, Skipper L, Hefferon T, Spalding JD, Garner J, Chen C, Maguire M, Corbett M, Zhou G, Paschall J, Ananiev V, Flicek P, Church DM. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. [PMC free article: PMC3531204] [PubMed: 23193291]
- 2.
- 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [PMC free article: PMC3498066] [PubMed: 23128226]